DEPARTMENT OF MOLECULAR PHYSIOLOGY
Outline
Department of Molecular Physiology constitutes of 3 Divisions (Division of Biophysics and Neurobiology, Division of Neurobiology and Bioinformatics, and Division of Nano-Structure Physiology) and is mainly involved in "Clarification of Function and Regulation of Bioactive Molecules", which is one of the five major projects settled in our institute. Also it is actively involved in the project "Development of integrated 4 dimensional imaging of brain structure and bioactive molecules".
Ion channels, receptors and G proteins play critical roles for the excitability and its regulation of neurons. We focus on these molecules which enable brain function. From the biophysical point of view, we study structure-function relationships, regulation mechanisms and dynamic structural rearrangements of ion channels and receptors. We also study the functional significance of specific features of ion channels and receptors in the brain function by making knock-in mice and by studying their abnormalities in the synaptic transmission and whole animal behavior. Specific themes of research projects currently running are as follows.
- Structure-function relationship and heteromultimeric subunit assembly of inwardly rectifying K+ channels.
- Molecular mechanisms and functional significance of the Ca2+/Gd3+ sensing function and multiple path signaling switching of the metabotropic glutamate receptor.
- Analysis of the dynamic structural rearrangements of ion channels and receptors by FRET measurement under evanescent field illumination.
- Changes of the voltage-sensor movement of KCNQ1 channel by assembly of KCNE subunit and analyses of the stoichiometry of assembly by single molecule imaging.
- Voltage-dependent gating of ATP receptor channel P2X2.
- Molecular identification and functional analysis of prestin complex, a motor protein of the outer hair cell.
- Identification of the molecular determinant of TRPA1 channel for the species dependent difference of the caffeine sensitivity.
- Molecular identification of the Cs+ permeable K+ channel in the cerebellar Purkinle cells activated by GABAB receptor stimulation.
- Analyses of the expression pattern and function of an orphan metabotropic receptor Prrt3.
- Functional analysis of RGS family, regulators of G protein signaling.
- Purification of recombinant proteins of P2X2 channel and Prestin toward single particle structure analysis.
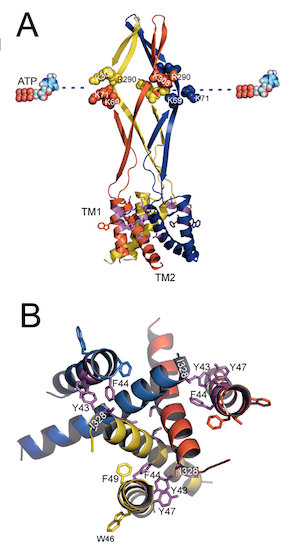
Fig. 1. Functional and structural identification of amino acid residues of the P2X2 receptor channel critical for the voltage- and [ATP]-dependent gating. (Keceli and Kubo, J. Physiol., 2009)
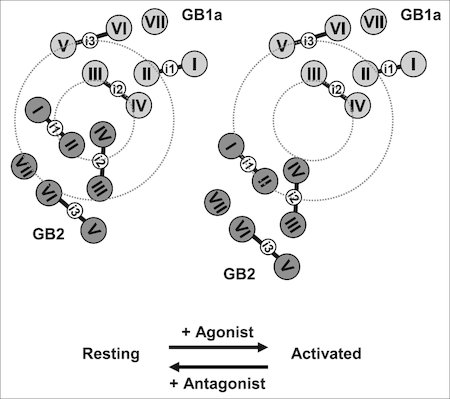
Fig. 2. Ligand-induced rearrangements of the GABAB receptor revealed by fluorescence resonance energy transfer (Matsushita, Nakata, Kubo and Tateyama, J. Biol. Chem., 2010)
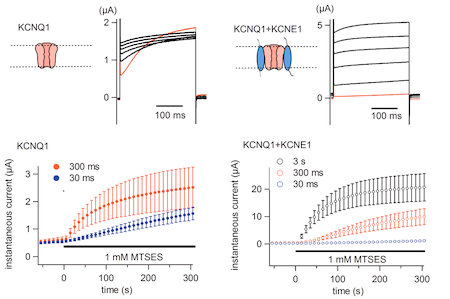
Fig. 3. KCNE1 and KCNE3 stabilize and/or slow voltage sensing S4 segment of KCNQ1 channel. (Nakajo and Kubo, J. Gen. Physiol., 2007)
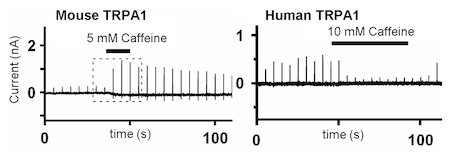
Fig. 4. Caffeine activates mouse TRPA1 channels but suppresses human TRPA1 channels. (Nagatomo and Kubo, Proc. Natl. Acad. Sci. USA, 2008)
Staff
KUBO, Yoshihiro, MD, PhD
1985 Graduated from University of Tokyo, Faculty of Medicine. 1989 Completed the doctoral course in Medical Science, University of Tokyo. 1989-2000 Researcher, Tokyo Metropolitan Institute for Neuroscience. (1991-1993: Post-doc, University of California, San Francisco). 2000 Professor, Tokyo Medical and Dental University Graduate School of Medicine. 2003 Professor, NIPS.
Specialty: Biophysics, Neurobiology
TATEYAMA, Michihiro, PhD
1990 Graduated from University of Tokyo, Faculty of Pharmacology. 1995 Completed the doctoral course in Pharmacology, University of Tokyo. 1995-2000 Assistant Professor, Juntendo University School of Medicine. 2000-2002 Research Fellow, Columbia University. 2002-2004 Research Fellow, CREST. 2004 Associate Professor, NIPS.
Specialty: Pharmacology, Physiology
NAKAJO, Koichi, PhD
1997 Graduated from University of Tokyo, College of Arts and Sciences. 2002 Completed the doctoral course in Life Science, University of Tokyo Graduate School of Arts and Sciences. 2002 Inoue Research Fellow. 2004 Research Fellow, NIPS. 2005 Assistant Professor, NIPS.
Specialty: Molecular Physiology, Biophysics
KECELI, Mehmet Batu, MD, PhD
1999 Graduated from Hacettepe University Faculty of Medicine (Turkey), 2001 Military Obligation, 2002 General Practitioner, Kocaeli Emniyet Hospital, 2003 Research Assistant, Hacettepe University Faculty of Medicine, 2004 Completed the master course, Hacettepe University, 2009 Completed the doctoral course, SOKENDAI.
Specialty: Molecular Physiology, Biophysics
During the course of formation of the mammalian central nervous system, neuroepithelial cells differentiate into various kinds of cells to make a fine three-dimensional network. Our goal is to understand genetic control over these processes. As a first step, we have cloned several genes that are specifically expressed in a certain type of brain cells and are investigating their role on cell fate determination. Neural cells are known to leave the ventricular zone after their commitment, and migrate towards destinations. While radial neuronal migration has been studied extensively in the developing cerebral and cerebellar cortices, mechanisms underlining tangential migration of neuronal andglial progenitors remains unclear. We are employing in ovo or in utero electroporation method to introduce exogenous genes in developing central nervous system, and studying mode and mechanisms of neural cell migration.
We are making use of hereditary mutant mice that exhibit abnormal development of the nervous system. We also use in situ hybridization and immunohistochemical technique to study cell lineages during development of the nervous system.
Neural stem cells, which are ultimate lineage precursors to all neurons and glia in the mammalian brain, are present not only in embryonic but also in adult brains, and contribute to adult neurogenesis. We are investigating molecular mechanisms underlying the generation, proliferation, maintenance, differentiation, and senescence of the neural stem cells, which will clarify their in vivo kinetics and function.
An automated system to analyze N-linked sugar chains was developed to study their biological roles during development and tumorigenesis.
New retroviral vectors are also constructed for efficient gene delivery, which will be used for cancer gene therapy.
A)
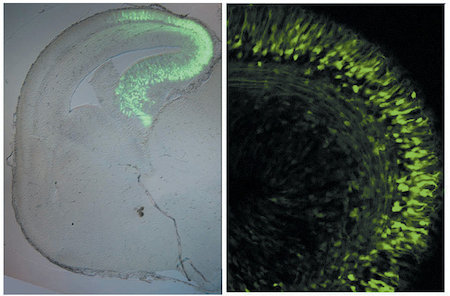
A) In utero electroporation was carried out for plasmid DNA transfer. Green fluorescent protein (GFP) expression vector was injected into lateral ventricle and electroporated in utero. The cells in the restricted region were observed to express GFP
B)
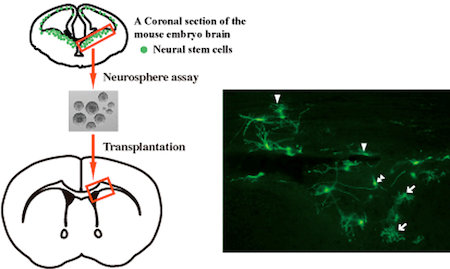
B) Transplantation of neural stem cells into the adult brain
Neural stem cell-derived neurospheres which are cultured from GFP+ mouse embryos are transplanted into mouse adult brains. Four weeks after the transplantation, GFP+ astrocytes (arrows), oligodendrocytes (arrow heads) and neurons (double arrow heads) are present in the adult brain.
C)
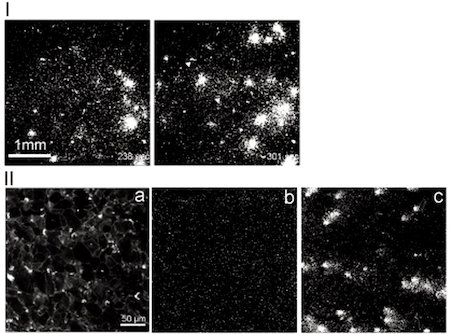
C) Imaging of ATP and glutamate release.
Ⅰ)ATP was released from astrycytes by hypotonic stimulation. ATP was released from astorocytes during about 100~200 sec.
Ⅱ)Glutamate optic sensor was immobilized onto membrane of astrocytes.
Glutamate was released from astrocytes by hypotomic stimulation (a). To visualize glutamate releasem the change in the fluorescent intensity was observed (b and c). Before hypotonic stiumulation, astrocytes did not release glutamate (b). After the stimulation, glutamate was released from astrocytes during around 10~20 sec (c).
Staff
IKENAKA, Kazuhiro, PhD
1975 Graduated from Faculty of Science, Osaka University. 1980 Graduated from the doctoral course at Osaka University, PhD. 1980 Instructor at Institute for Protein Research, Osaka University. 1991 Associate Professor at Institute for Protein Research, Osaka University. 1992 Professor, NIPS.
Specialty: Molecular Neurobiology
HITOSHI, Seiji, MD, PhD
1988 Graduated from Faculty of Medicine, University of Tokyo. MD. 1993 Board-certified neurologist by Japanese Society for Neurology. 1997 PhD from Graduate School of Medicine, University of Tokyo. 1997 Special Postdoctoral Researcher at the Institute of Physical and Chemical Research (RIKEN). 1999 Postdoctoral Fellow at University of Toronto. 2003 Assistant Professor at University of Tokyo. 2003 Associate Professor at NIPS.
Specialty: Developmental Neurobiology, Neurology
TANAKA, Kenji, MD, PhD
1997 Graduated from Keio University, School of Medicine. 1997-1999 Resident in Department of Neuropsychiatry, Keio University, School of Medicine. 2003 Completed the doctoral course in Keio University. 2003 Research Associate, NIPS. 2004 Assistant Professor, NIPS.
Specialty: Neurochemistry, Biological psychiatry
YOSHIMURA, Takeshi, PhD
2000 Graduated from Kyushu University, School of Agriculture. 2002 Graduated from NAIST, Graduate School of Biological Sciences. 2005 Graduated from Nagoya University, Graduate School of Medicine, PhD. 2005-2008 Postdoctoral Fellow, Nagoya University. 2008 Assistant Professor, NIPS.
Specialty: Molecular Biology, Neuroscience
INAMURA, Naoko, PhD
Graduated from Kyoto Institute of Technology, Department of Applied Biology. Graduated from the master course in Kobe University, School of Science and Technology. Graduated from the doctoral course in Osaka University, School of Science, PhD. Postdoctoral Fellow, NIPS.
Specialty: Neural Development
A novel methodology, when it is very informative, gives an aid to the opening of a novel scientific field. For example, MRI originally born from NMR in chemistry and primarily developed for diogonoes has outgrown to cover almost all medical sciences. We call such a productive innovation, emerged from an old regime but creative to a new field, as a strategic methodology. Integration of biosciences might bring about such a difficulty that a simple sum of constituent disciplines never makes a good start. Fusion of different disciplines can be encouraged by novel breakthroughs in methodology. The expected are new methods for three-dimensional structural analysis of single biological molecules and in situ functional observation of complex biological systems.
This laboratory works on methodological themes by relying on the technical breakthrough of imaging methods such as electron microscopy.
(1) Development and application of electron-phase microscopy: Different kinds of phase observation schemes have been developed including the novel optical principle for the reconstruction of complex wavefunctions. They are expected to enhance the contrast of biological samples which is inherently very poor in electron microscopy.
Applications are:
i) direct visualization of protein molecules or cytoskeltons in the in vivo state of cells and tissues,
ii) structural and functional analyses of membrane proteins and viruses with the aid of single particle analysis,
iii) photon-electron hybrid electron microscopy to visualize intact neurons at a high resolution.
(2) Biological transports: Transcellular and paracellurar mechanisms for transport of water, electrolytes, and substrates are investigated by laying much emphasis on molecular mechanism of exocytosis and energy supply for transport in the exocrine glands.
(3) Sorting in the endocytic pathway: The endocytic pathway functions as a sorting station for molecules that are destined either for lysosomes (a degrative pathway) or for recycling pathways, thereby determining the fate of endomembrane molecules. The physiological roles and the mechanisms of sorting in the endocytic pathway are investigated.
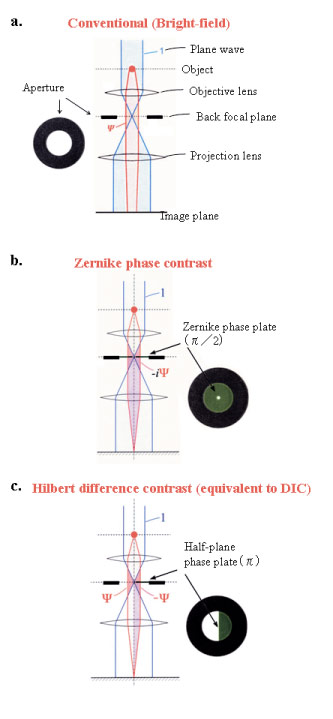
Fig. 1 Three kinds of schemes for electron-phase microscopy.
a. Conventional (bright-field) method to enhance the image contrast at the expense of a deterioration of the spatial resolution by defocusing.
b. Zernike phase-contrast method to enhance the image contrast under the just focus condition by inserting a Zernike phase plate to the objective back-focal plane.
c. Hilbert differential method to obtain phase contrast images similar to light-microscopic DIC (Differential-Interference-Contrast) images by inserting a half-plane p phase plate to the objective back-focal plane.
(adapted from Biophs. Rev 1 (2009)37)
Fig. 2 Two kinds of electron-phase microscope models.
a. 300kV analytical cryoelectron microscope (equipped with a FEG, a He-stage and a energy filter) with phase plates.
b. 200kV tomography-dedicated cryoelectron microscope (equipped with a FEG, a tilting N2- stage and a energy-filter) with phase plates.
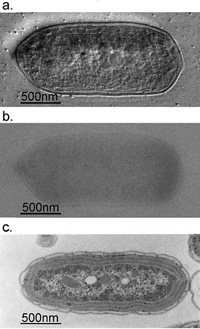
Fig. 3 Comparison of 300kV and 100kV TEM images for ice-embedded and plastic-embedded cyanobacterial cells.
a. A 300kV Hilbert differential image for an ice-embedded cyanobacterial whole cell, which holds a resolution sufficient enough to identify subcellular structures down to 2nm.
b. A 300kV conventional image shot for the same sample as shown in a., of which low contrast makes it difficult to identify subcellular structures.
c. A 100kV conventional image for a plastic-embedded and thin-sectioned cyanobacterial cell, which was prepared with a chemical fixation, dehydration and a heavy metal staining. Due to the harsh and lengthy chemical treatments, subcellular structures are heavily damaged making their morphological preservation hard.
(taken from Kaneko et al., J. Electro. Microsc. 54 (2005) 79)
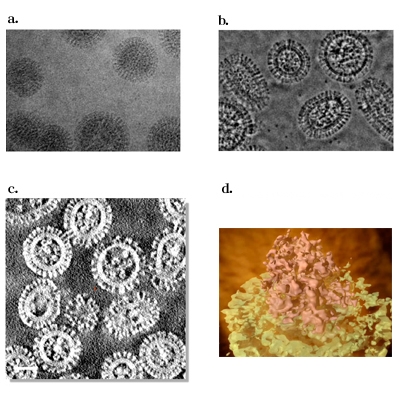
Fig. 4 Electron micrograms and tomograms of Influenza A virus.
a. A conventional microgram (300kV).
b. A Zernike phase-contrast microgram (300kV)
c. A slice of Zernike phase-contrast tomogram (200kV)
d. A 3D image of influenza genome reconstituted from Zernike phase contrast tomogram (200kV)
(a and b taken from Yamaguchi et al., J. Struct. Biol., 162 (2008) 271, c and d taken from Danev et al., unpublished. 3D graphics shown in d. is creditted to Takaaki Takeda of Imaging Science Research group, NINS)
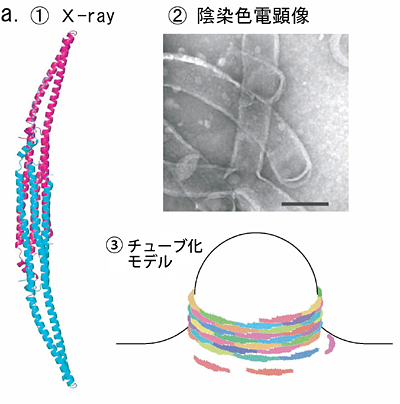
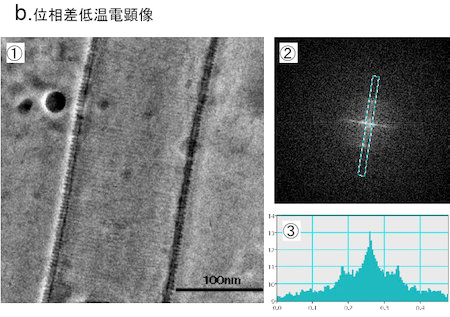
Fig.5 Phase contrast cryo-TEM fills the gap beetweem X-ray crystallography and conventional TEM observation
a. Lipid interacting protein, PCH, has a domain called EFC-domain assumed to be responsible for the liposome tubulation. X-ray crystallography revealed a thread structure (①) and conventioned TEM observation with negative staining (②) showed a fixed size tubulation induced by the EFC-domain. From the two results a model as shown in ③ has been proposed.
b. The proposed model has been visually proven with the phase contrast cryo-TEM as shown in ①. The Fourier analysis (②, ③) tells us the spacing of adjacently wound EFC-domain polymers being 4nm, which is just expected from the X-ray structure shown in a-① (taken from Shimada et al., Cell , 129 (2007) 176).
Staff
NAGAYAMA, Kuniaki, PhD
1968 Graduated from University of Tokyo. 1973 Completed the doctoral course in Science, University of Tokyo. 1974 Research Associate, University of Tokyo. 1984 Director, Biometrology Lab, JEOL Ltd. 1990 Project Leader, Nagayama Protein Array Project, ERATO, JRDC. 1993 Professor, The University of Tokyo. 1997 Professor, NIPS. 2001 Professor, Okazaki Institute for Integrative Bioscience (OIB).
Speciality: Biophysics, Electron Microscopy
MURAKAMI, Masataka, MB, M.D.
1976 Graduated from Kyoto Prefectural University of Medicine. 1976 Research Associate, Osaka Medical College. 1981 Doctor of Medicine in Physiology of Osaka Medical College. 1983 Postdoctorial Fellow, Department of Physiology, University of Sydney. 1985 Associate Professor, NIPS. 2003 Associate Professor, OIB (Seconded from NIPS).
Speciality: Physiology of exocrine glands, Energy metabolism and transport of electrolyte and water, Paracellular Transport
OHASHI, Masato, PhD
1986 Graduated from Kyoto University, Faculty of Science. 1992 Completed the doctoral course in Science, Kyoto University. 1992 Postdoctoral Fellow, Department of Neurobiology, University of Heidelberg. 1996 Assistant Professor, NIPS. 2003 Assistant Professor, OIB.
Speciality: Cell Biology
DANEV, Radostin, PhD
1997 Graduated from Faculty of Physics, Sofia University, Sofia, Bulgaria. 2001 Completed the doctoral course in Science, The Graduate University for Advanced Studies, NIPS, Okazaki. 2001 Postdoctoral Fellow, NIPS. 2002 Research fellow, OIB. 2006 JST Assistant Professor, OIB. 2008 Assistant Professor, OIB.
Speciality: Solid State Physics, Electron Microscopy
HOSOGI, Naoki
2003 Graduated from University of Kobe, Faculty of Agriculture. 2008 Completed the doctoral course in Bioresource and Agrobiosciences, University of Kobe. 2008 Postdoctoral Fellow, OIB.
Speciality: Phytopathology, Plant Cell Biology
KAYAMA, Yoko
1998 Graduated from Shinshu University, Faculty of Engineering. 2007 Completed the doctoral course in Science and Technology, Meijo University. 2008 Postdoctoral Fellow, OIB.
Speciality: Materials science
FUKUDA, Yoshiyuki
2004 Graduated from Kanagawa University, Faculty of science. 2009 Completed the doctoral course in School of Life science, the Graduate University for Advanced Studies. 2009 Postdoctoral Fellow, OIB.
Speciality: Neuroanatomy















