Novel structural determinants for the slow deactivation of the hERG channel
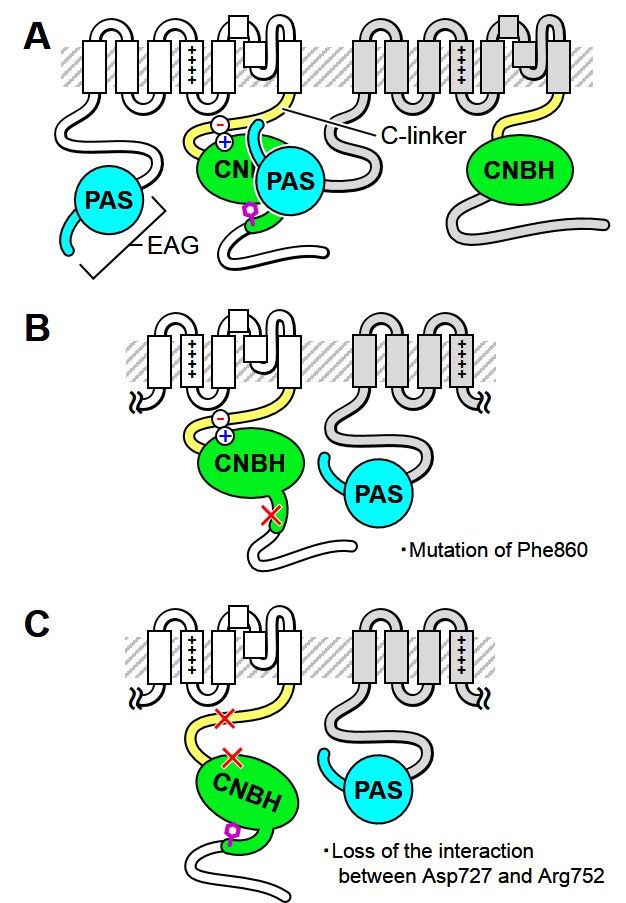
The human ether-a-go-go related gene (hERG) channel, a member of the voltage-gated K+ channel family, plays an important role in the repolarization phase of the action potential in human cardiac myocytes, as IKr, and shows characteristic slow deactivation, which is needed an interaction between the N-terminal cytoplasmic ether-a-go-go (EAG) domain (Figure A) and the C-terminal cytoplasmic cyclic nucleotide (CN) binding homology (CNBH) domain (Figure A). Loss of function of the hERG channel by genetic abnormality or drug application is causative of Long QT syndrome, which induces ventricular arrhythmia or sudden death.
In this study, we analyzed the effects of mutations in the CNBH domain and its upstream C-linker domain, the detailed mechanisms of both domains are unknown, on slow deactivation and the interaction between the EAG and CNBH domains, by electrophysiological and FRET analyses using Xenopus oocyte and HEK293T cell expression systems. The mutations of Phe860 (Figure B), which is reported to fill the CN binding pocket as an intrinsic ligand, and Asp727 & Arg752 (Figure C), which forms a newly identified salt bridge pair between C-linker and CNBH domains, accelerated deactivation and also decreased the interaction between the EAG and CNBH domains. Furthermore, the FRET analysis shows the voltage-dependent changes between the two domains were not detected.
The results suggest that the CNBH domain (and C-linker domain) contributes to slow deactivation of the hERG channel by a mechanism involving the EAG domain.