We investigate structures of biomolecules, biomacromolecular complexes, and cellular architecture using electron microscopy, and reveal a relationship between the structure and the function.

We have succeeded in elucidating the fiber structure of the familial Tottori type (D7N mutation) of amyloid beta (Aβ), the causative agent of Alzheimer's disease, through experiments utilizing the microgravity environment of the International Space Station.
Yagi-Utsumi M, Yanaka S, Burton-Smith RN, Song C, Ganser C, Yamazaki C, Kasahara H, Shimazu T, Uchihashi T, Murata K, Kato K (2025) Microgravity-Assisted Exploration of the Conformational Space of Amyloid β Affected by Tottori-Type Familial Mutation D7N. ACS Chem Neurosci 16(14) 2682-2690. WEB
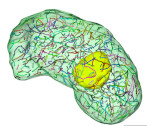
We used volume EM to perform a detailed analysis of the nuclear structure of the dinoflagellate Oxyris marina, revealing for the first time that this species has nearly 400 chromosomes (dinokaryons) unique to this species.
Fukuda Y, Suzaki T, Murata K, Chihong S (2025) Novel ultrastructural features of the nucleus of the ancestral dinoflagellate Oxyrrhis marina as revealed by freeze substitution fixation. Front Protistol 3 1512258. WEB
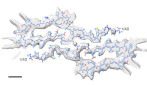
Cryo-EM structural analysis of wild-type Aβ(1-40) fibers formed on Earth in a manner similar to that in the microgravity environment of the ISS revealed a novel J-shaped structure.
Burton-Smith RN, Yagi-Utsumi M, Yanaka S, Song C, Murata K, Kato K (2025) Elucidating the Unique J-Shaped Protomer Structure of Amyloid-β(1-40) Fibril with Cryo-Electron Microscopy. Int J Mol Sci 26(3) 1179. WEB
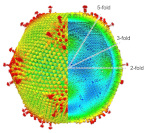
The capsid structure of Medusavirus was analyzed at 7 Å resolution using cryo-EM, revealing structural changes in the capsid that accompany the particle formation process.
Watanabe R, Song C, Takemura T, Murata K* (2024) Subnanometer structure of medusavirus capsid during maturation using cryo-electron microscopy. J Virol 98(9), e0043624. WEB
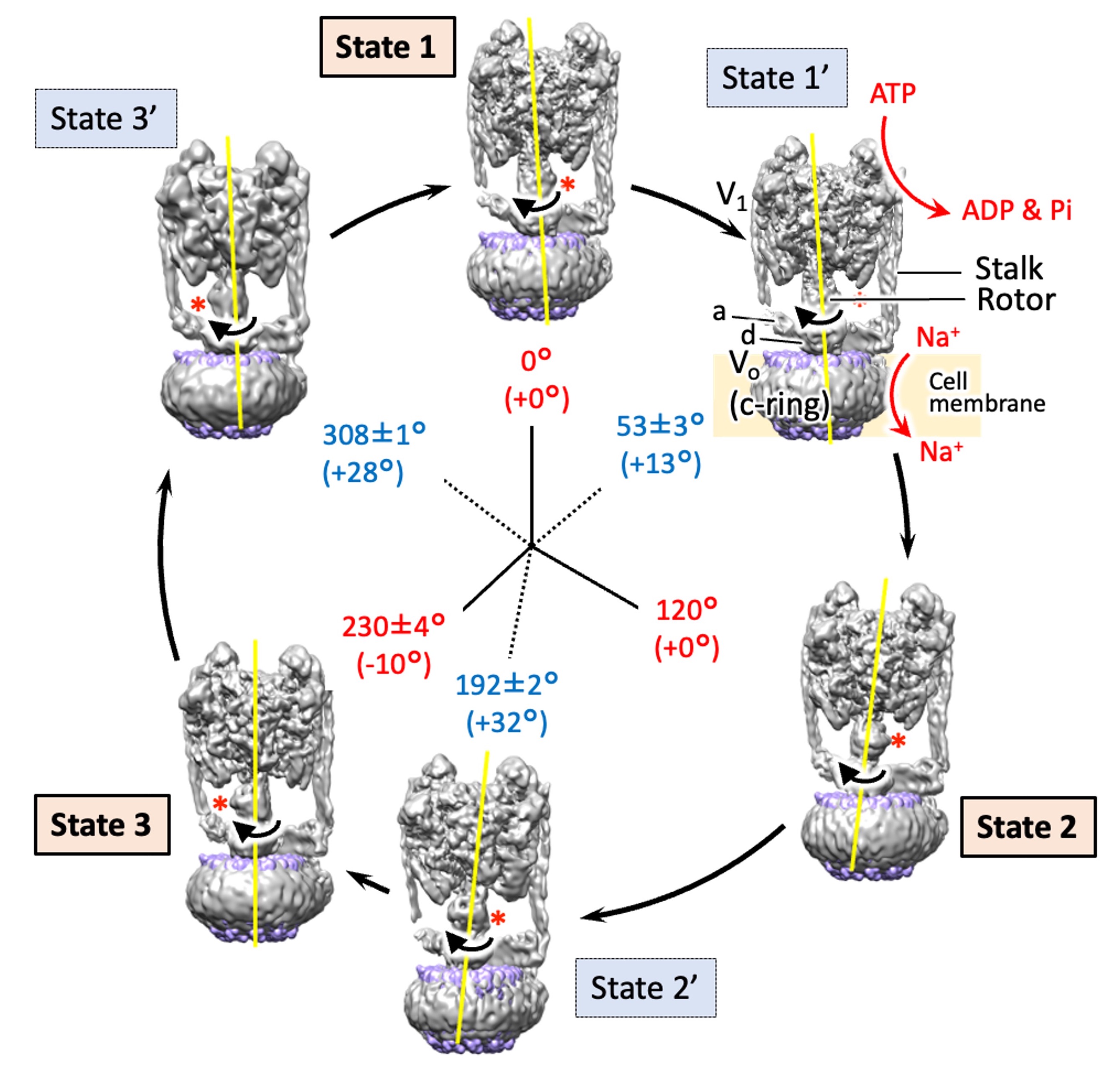
The structures of six states of a rotary sodium ion pump are revealed using cryo-electron microscopy,elucidating the branched mechanism by which sodium is transported by hydrolyzing ATP. .
Raymond N. Burton Smith, Chihong Song, Hiroshi Ueno, Takeshi Murata, Ryota Iino, Kazuyoshi Murata* (2023) Six states of Enterococcus hirae V-type ATPase reveals non-uniform rotor rotation during turnover. Comm Bio 6 755. WEB
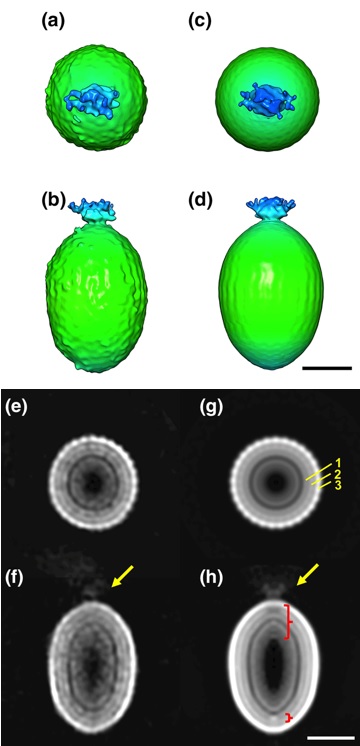
The detailed particle structure of Tanaivirus was revealed using cryo-electron microscopy, revealing the dynamic structural changes of the capsid that occur during infection.
Kenta Okamoto*, Chihong Song, Han Wang, Miako Sakaguchi, Christina Chalkiadaki, Naoyuki MIyazaki, Takeshi Nabeshima, Kouichi Morita*, Shingo Inoue, and Kazuyoshi Murata (2023) Structure and its transformation of elliptical nege-like virus Tanay virus. J Gen Vir 104(6),001863. WEB
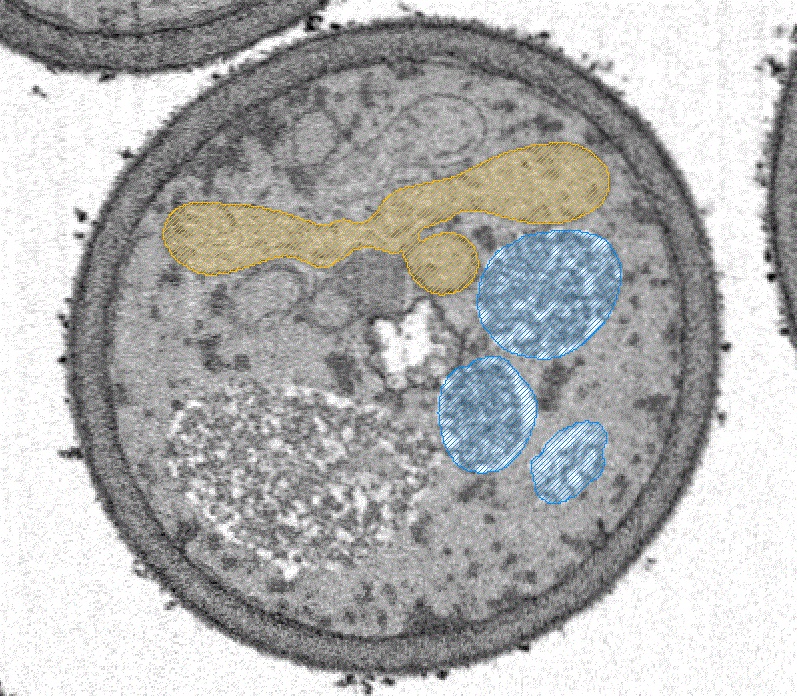
New factor of mitochondrial fission in mitophagy was discovered and the process was visualized by 3D-SEM.
Tomoyuki Fukuda#, Kentaro Furukawa#, Tatsuro Maruyama#, Shun-Ichi Yamashita, Daisuke Noshiro, Chihong Song, Yuta Ogasawara, Kentaro Okuyama, Jahangir Md. Alam, Manabu Hayatsu, Tetsu Saigusa, Keiichi Inoue, Kazuho Ikeda, Akira Takai, Lin Chen, Vikramjit Lahiri, Yasushi Okada, Shinsuke Shibata, Kazuyoshi Murata, Daniel J. Klionsky, Nobuo N. Noda*, Tomotake Kanki* (2023) The mitochondrial intermembrane space protein mitofissin drives mitochondrial fission required for mitophagy. Mol Cell 83(12),2045-2058. WEB
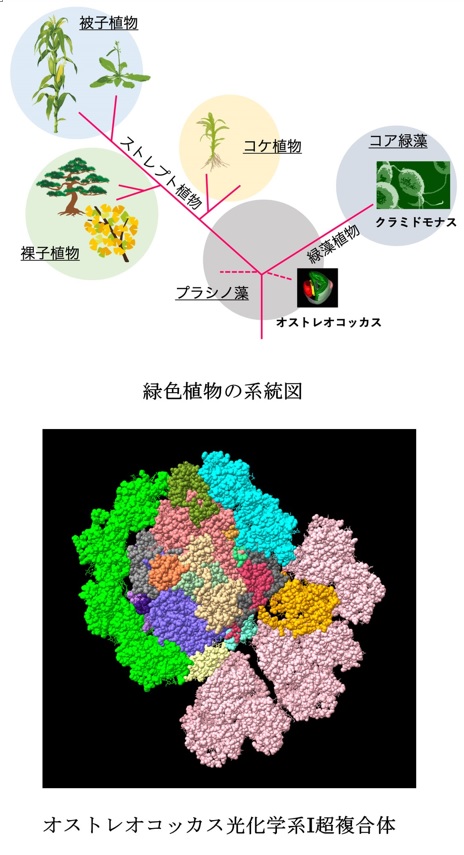
The unique structure of acient type PSI in Ostreococcal was revealed using cryo-electron microscopy.
Asako Ishii, Jianyu Shan, Xin Sheng, Eunchul Kim, Akimasa Watanabe, Makio Yokono, Chiyo Noda, Chihong Song, Kazuyoshi Murata, Zhenfeng Liu, Jun Minagawa (2023) The photosystem I supercomplex from a primordial green alga Ostreococcus tauri harbors three light-harvesting complex trimers. eLife 84488. WEB
Structure of a giantvirus, tokyovirus, was revealed at 7.7Å resolution by high-voltage cryo-EM single particle analysis.
Chihara A, Burton-Smith RN, Kajimura N, Mitsuoka K, Okamoto K, Song C, Murata K (2022) A novel capsid protein network allows the characteristic inner membrane structure of Marseilleviridae giant viruses. Sci Rep 12(1):21428. WEB
Hydrated structures of nano-composite microgels produced with different components were revealed by cryo-electron tomography.
Nishizawa Y, Watanabe T, Noguchi T, Takizawa M, Song C, Murata K, Minato H, Suzuki D (2022) Durable gelfoams stabilized by compressible nanocomposite microgels. Chem Commun (Camb) 58, 12927-12930. WEB
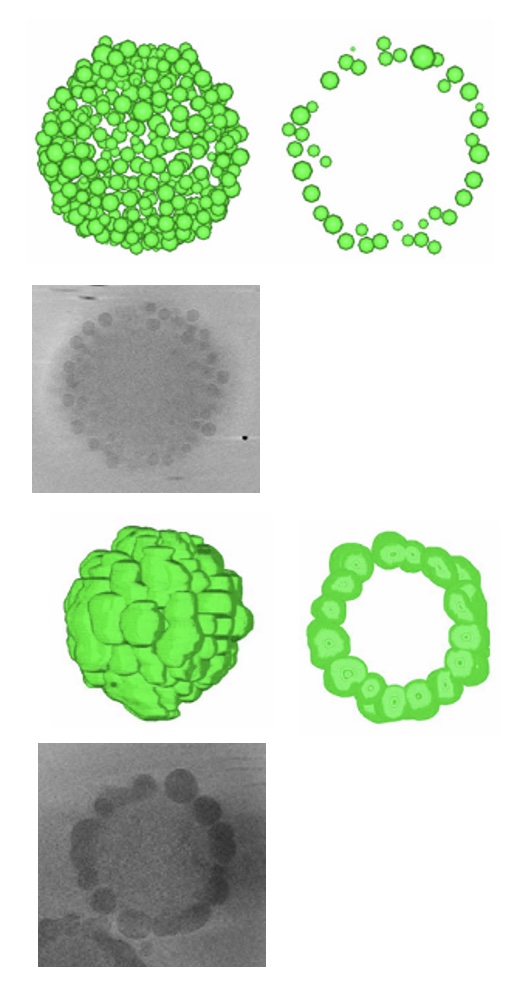
The structure of the sapovirus shell (capsid), one of the causative viruses of mass diarrhea, was clarified for the first time in the world at the atomic level.
Miyazaki N, Song C, Oka T, Miki M, Murakami K, Iwasaki K, Katayama K, Murata K* (2022) Atomic structure of human sapovirus capsid by single particle cryo-electron microscopy revealed a unique capsid protein conformation in caliciviruses. J Virol 96(9), e0029822. WEB
The neural circuit (connectomics) of the brain region where the light information entering from the compound eye of the swallowtail butterfly is initially processed has been clarified.
Matsushita A, Stewart F, Ilić M, Chen PJ, Wakita D, Miyazaki N, Murata K, Kinoshita M, Belusic G, Arikawa K (2022) Connectome of the lamina reveals the circuit for early color processing in the visual pathway of a butterfly. Cur biology 32(10):2291.e3, 00506-1. WEB
Medusavirus, a giant virus, shows a unique maturation process in host amoeba.
Watanabe R, Song C, Kayama Y, Takemura M, Murata K* (2022) Particle morphology of Medusavirus inside and outside cells reveals new maturation process of giant virus. J Viol 96(7), e0185321. WEB
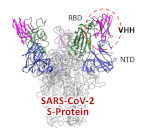
A neutralized VHH antibody for SARS-CoV2 was developed and its binding model was revealed by cryo-EM.
Haga K, Takai-Todaka R, Matsumura Y, Song C, Takano T, Tojo T, Nagami A, Ishida Y, Masaki H, Tsuchiya M, Ebisudani T, Sugimoto S, Sato T, Yasuda H, Fukunaga K, Sawada A, Nemoto N, Murata K, Morimoto T, Katayama K. (2021) Nasal delivery of single-domain antibody improves symptoms of SARS-CoV-2 infection in an animal model. PLoS Pathog 17(10):e1009542. doi: 10.1371/journal.ppat.1009542. WEB
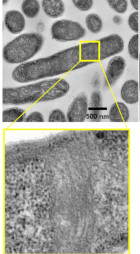
CAHS protein in targigrade was revealed to form a fiber in desiccated condition.
Yagi-Utsumi M, Aoki K, Watanabe H, Song C, Nishimura S, Satoh T, Yanaka S, Ganser C, Tanaka S, Schnapka V, Goh EW, Furutani Y, Murata K, Uchihashi T, Arakawa K, Kato K (2021) Desiccation-induced fibrous condensation of CAHS protein from an anhydrobiotic tardigrade. Sci Rep 11(1), 21328. doi: 10.1038/s41598-021-00724-6. WEB
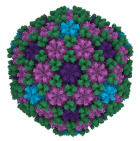
Helicobacter pylori bacteriophage exhibits a capsid structure to avoid digestion in stomach.
Kamiya R, Uchiyama J, Matsuzaki S, Murata K, Iwasaki K, Miyazaki N (2021) Acid-stable capsid structure of Helicobacter pylori bacteriophage KHP30 by single-particle cryo-electron microscopy. Structure 30(2):300-312.e3. doi: 10.1016/j.str.2021.09.001 WEB
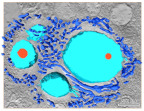
Cotonvirus, a giant virus descovered in Japan uses Golgi apparatus for its virion factory.
Takahashi H, Fukaya S, Song C, Murata K, Takemura M (2021) Cotonvirus japonicus using Golgi apparatus of host cells for its virion factory phylogenetically links tailed tupanvirus and icosahedral mimivirus.J Virol 95(18),e0091921. doi: 10.1128/JVI.00919-21. WEB
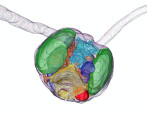
A phytoplankton isodated in north see was found to produces oil.
Harada N, Hirose Y, Song C, Kurita H, Sato M, Onodera J, Murata K, Itoh F (2021) A novel characteristic of a phytoplankton as a potential source of straight-chain alkanes.Sci Rep 11(1), 14190. WEB

The state transition structure of PS-I was revealed in Chlamydomonas.
Pan X , Tokutsu R, Li A, Takizawa K, Song C, Murata K, Yamasaki T, Liu Z, Minagawa J, Li M (2021) Structural basis of LhcbM5-mediated state transitions in green algae.Nature plant 7(8), 1119-1131. WEB

The double ring consisting of the proteasome α7 subunit has large fluctuations, which provide hints for the formation of the proteasome complex.
Song C, Satoh T, Sekiguchi T, Kato K*, Murata K* (2021) Structural fluctuations of the human proteasome α7 homo-tetradecamer double ring imply the proteasomal α-ring assembly mechanism. Int J Mol Sci 22(9), 4519. WEB
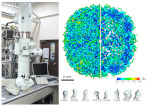
Sub-3 Å structure of apoferritin was achieved by a multipurpose TEM with a side entry cryoholder.
Kayama Y, Burton-Smith RN, Song C, Terahara N, Kato T, Murata K* (2021) Below 3 Å structure of apoferritin using a multipurpose TEM with a side entry cryoholder.Sci Rep 11, 8395. WEB
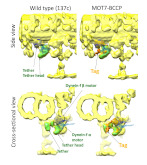
Location of the protein that controls ciliary movement by light was revealed in cilia.
Kutomi O, Yamamoto R, Hirose K, Mizuno K, Nakagiri Y, Imai H, Noga A, Obbineni JM, Zimmermann N, Nakajima M, Shibata D, Shibata M, Shiba K, Kita M, Kigoshi H, Tanaka Y, Yamasaki Y, Asahina Y, Song C, Nomura M, Nomura M, Nakajima A, Nakachi M, Yamada L, Nakazawa S, Sawada H, Murata K, Mitsuoka K, Ishikawa T, Wakabayashi KI, Kon T, Inaba K (2021) A dynein-associated photoreceptor protein prevents ciliary acclimation to blue light. Sci Adv 7, eabf3621. WEB
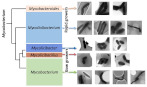
Five newly proposed genera of Mycobacterium tuberculosis exhibited different morphological characteristics.
Yamada H, Chikamatsu K, Aono A, Murata K, Miyazaki N, Kayama Y, Bhatt A, Fujiwara N, Maeda S, Mitarai S (2020) Fundamental cell morphologies of the species in the novel five genera robustly correlate with new classification in family Mycobacteriaceae. Front Microbiol 11, 562395. WEB

A new phase contrast STEM method using a Fresnel zone plate was developed, dramatically improving the image contrast of an electron microscope.
Tomita M, Nagatani Y, Murata K & Momose A (2020) Enhancement of low-spatial-frequency components by a new phase-contrast STEM using a probe formed with an amplitude Fresnel zone plate. Ultramicroscopy, 218, 113089. WEB
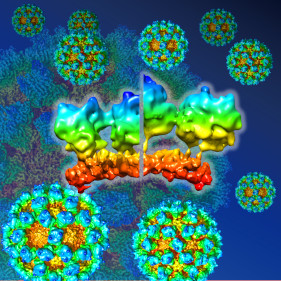
It was discovered that the norovirus could change its structure to allow infection.
Song C, Takai-Todaka R, Miki M, Haga K, Fujimoto A, Ishiyama R, Oikawa K, Yokoyama M, Miyazaki N, Iwasaki K, Murakami K, Katayama K* & Murata K* (2020) Dynamic rotation of the protruding domain enhances the infectivity of norovirus. PLOS Pathogens, 16(7), e1008619. WEB
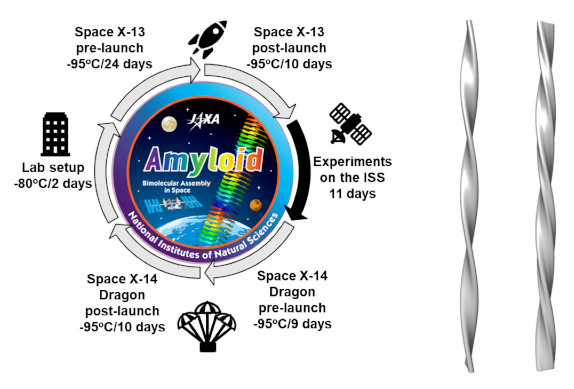
It was discovered that in a microgravity environment, unique forms of amyloid fibrils are formed. -An experiment at International Space Station "Kibo"-
Yagi-Utsumi M, Yanaka S, Song C, Satoh T, Yamazaki C, Kasahara H, Shimazu T, Murata K, Kato K (2020) Characterization of amyloid β fibril formation under microgravity conditions. Microgravity 6, 17. WEB
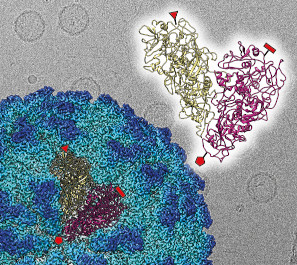
The functional evolution of RNA virus was clarified from the change of particle structure.
Okamoto K, Ferreira RJ, Larsson DSD, Maia FRNC, Isawa H, Sawabe K, Murata K, Hajdu J, Iwasaki K, Kasson PM, Miyazaki N (2020) Acquired functional capsid structures in metazoan totivirus-like dsRNA virus. Structure 28, 1-9. WEB
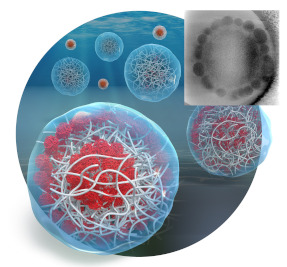
First preparation of multilayer nanocomposite hydrogel particles and their direct observation by cryo-electron tomography.
Watanabe T, Nishizawa Y, Minato H, Song C, Murata K, Suzuki D (2020) Hydrophobic monomers recognize microenvironments in hydrogel microspheres during free radical seeded emulsion polymerization. Angewandte Chemie Int Ed 59, 1-6. WEB
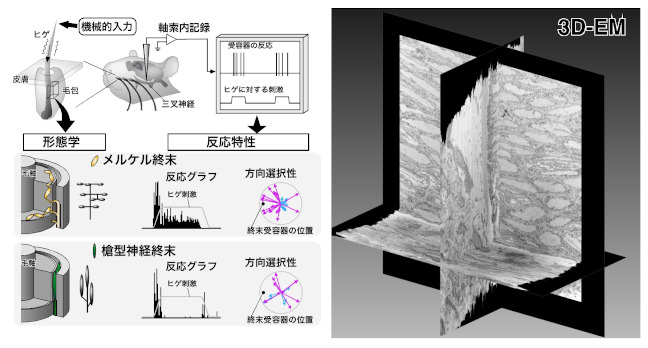
It was able to conduct detailed tactile experiments by targeting the rat vibrissal sensory system, and was clarify the role of nerve ending receptors in converting tactile information into neural activity.
Furuta T, Bush NE, Yang AET, Ebara S, Miyazaki N, Murata K, Hirai D, Shibata K & Hartmann MJZ (2020) The cellular and mechanical basis for response characteristics of identified primary afferents in the rat vibrissal system. Curr Biol 30, 815-826.e5. WEB
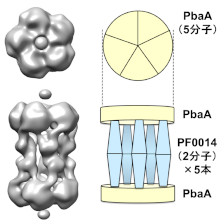
It was discovered a molecular building such as "tholos" formed by a hyperthermophilic archaeal protein.
Yagi-Utsumi M, Sikdar A, Song C, Park J, Inoue R, Watanabe H, Burton-Smith R.N., Kozai T, Suzuki T, Kodama A, Ishii K, Yagi H, Satoh T, Uchiyama S, Uchihashi T, Joo K, Lee J, Sugiyama M, Murata K & Kato K (2020) Supramolecular tholos-like architecture constituted by archaeal proteins without functional annotation. Sci Rep 10, 1540. WEB
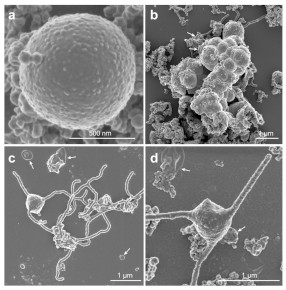
It was succeeded in culturing microorganisms most closely related to ancestors of eukaryotes, including humans, from deep-sea sediments. Cultivated microorganisms belong to a microbial strain called Archia, which grows depending on symbiosis with other microorganisms, and has genes that have been unique to eukaryotes (for example, genes that make actin and ubiquitin).
Imachi H, Nobu MK, Nakahara N, Morono Y, Ogawara M, Takaki Y, Takano Y, Uematsu K, Ikuta T, Ito M, Matsui Y, Miyazaki M, Murata K, Saito Y, Sakai S, Song C, Tasumi E, Yamanaka Y, Yamaguchi T, Tamaki H & Takai K (2020) Isolation of an archaeon at the prokaryote-eukaryote interface. Nature 577, 519–52515. WEB

>The three-dimensional structure of the photosystem II-light harvesting supercomposite of Chlamydomonas which is a huge light harvesting machine with a molecular weight of 1.66M was determined.
Sheng X, Watanabe A, Li A, Kim E, Song C, Murata K, Mr. Song D, Minagawa J & Liu Z (2019) Structural insight into light harvesting for photosystem II in green algae. Nature Plants 5, 1320–1330. WEB

In this study, we directly recorded the image of each nanoparticle using an electron microscope and reveal its size and particle size distribution accurately. It strongly contributes to the practical application of the multi-color total internal reflection dark-field microscope.
Ando, J, Nakamura A, Yamamoto M, Song C, Murata K & Iino R (2019) Multi-color high-speed tracking of single biomolecules with silver, gold, silver-gold alloy nanoparticles. ACS Photonics 6(11), 2870-2883. WEB
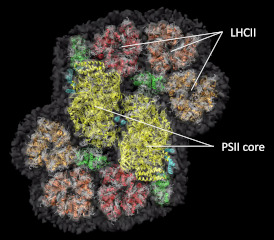
he structure of the giant photosynthetic membrane protein complex of Chlamydomonas was analyzed with a cryo-electron microscope at 5.6 Å resolution.
Burton-Smith, RN, Watanabe A, Tokutsu R, Song C, Murata K & Minagawa J (2019) Structural determination of the large photosystem II–light harvesting complex II supercomplex of Chlamydomonas reinhardtii using non-ionic amphipol. J Biol Chem 294(41), 15003–15013. WEB
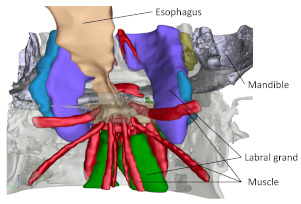
The state morphology of feeding adaptation and internal organ evolution was investigated in pelagic copepods by using SBF-SEM, and the relationship between environmental adaptation and integrated individual evolution of the components was clarified.
Kaji T, Song C, Murata K, Nonaka S, Ogawa, K, Kondo Y, Ohtsuka S & Palmer R (2019) Evolutionary transformation of mouthparts from particle-feeding to piercing carnivory in Viper copepods: 3D analyses of a key innovation using advanced imaging techniques. Front Zool 16, 35. WEB
It was clarified that subunits of proteolytic enzyme complex proteasome form various complexes depending on the combination.
Sekiguchi T, Satoh T, Kurimoto E, Song C, Kozai T, Watanabe H, Ishii K, Yagi H, Yanaka S, Uchiyama S, Uchihashi T, Murata K & Kato K (2019) Mutational and combinatorial control of self-assembling and disassembling of human proteasome α-subunits. Int J Mol Sci 20(9), 2308. WEB
It was discovered that cyanobacteria has a new chromatic acclimation processes regulating phycoerythrocyanin.
Hirose Y, Chihong S, Watanabe M, Yonekawa C, Murata K, Ikeuchi M & Eki T (2019) Diverse chromatic acclimation regulating phycoerythrocyanin and rod-shaped phycobilisome in cyanobacteria. Mol Plant 12(5), 715-725. WEB

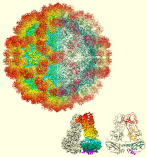
A novel giant virus with a full set of histone genes possessed by eukaryotes was discovered in Japanese hot spring. The unique capsid structure was revealed by cryo-electron microscopic single particle analysis.
Yoshikawa G, Blanc-Mathieu R, Song C, Kayama Y, Mochizuki T, Murata K, Ogata H & Takemura M (2019) Medusavirus, a novel large DNA virus discovered from hot spring water. J Virol 93(8), e02130-18. WEB
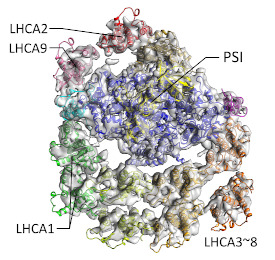
The whole structure of the super-giant photosynthetic complex PSI-LHCI of the green alga Chlamydomonas was clarified by cryo-.
Kubota-Kawai H, Burton-Smith RN, Tokutsu R, Song C, Akimoto S, Yokono M, Ueno Y, Kim E, Watanabe A, Murata K & Minagawa J (2019) Ten antenna proteins are associated with the core in the supramolecular organization of the photosystem I supercomplex in Chlamydomonas reinhardtii. J Biol Chem 294(12), 4304-4314, WEB
The process of the capsid formation of the rice dwarf virus was clarified using a phase-contrast cryo-electron microscope.
Nakamichi Y, Miyazaki N, Tsutsumi K, Higashiura A, Narita H, Shimizu T, Uehara-Ichiki T, Omura T, Murata K Nakagawa A (2019) An assembly intermediate structure of Rice dwarf virus reveals a hierarchical outer capsid shell assembly mechanism. Structure 27, 1-10. WEB

The molecular mechanism that metal nickel inhibits p53 protein complex formation was investigated by electron microscopy.
Kim YJ, Lee YJ, Kim HJ, Kim HS, Kang M-S, Lee S-K, Park MK, Murata K, Kim HL & Seo YR (2018) A molecular mechanism of nickel (II): reduction of nucleotide excision repair activity by structural and functional disruption of p53. Carcinogenesis 39, 1157–1164.WEB
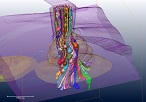
The entire structure of Enterococcal V-ATPase was revealed using a Zernike phase-contrast cryo-electron microscope.
Tsunoda J, Song C, Imai FL, Takagi J, Ueno H, Murata T, Iino R & Murata K (2018) Off-axis rotor in Enterococcus hirae V-ATPase visualized by Zernike phase plate single-particle cryo-electron microscopy. Sci Rep 8, 15632, WEB
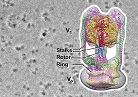
3D structure of the ant tactile nerve system was investigated using SBF-SEM.
Takeichi Y, Uebi T, Miyazaki N, Murata K, Yasuyama K, Inoue K, Suzaki T, Kubo H, Kajimura N, Takano J, Omori T, Yoshimura R, Endo Y, Hojo MK, Takaya E, Kurihara S, Tatsuta K, Ozaki K & Ozaki M (2018) Putative Neural Network within an Olfactory Sensory Unit for Nestmate and Non-nestmate Discrimination in the Japanese Carpenter Ant: The Ultrastructures and Mathematical Simulation. Front Cell Neurosci, 12, 310. WEB

The shape of the synthetic hydrogel particles was investigated by cryo-electron microscope tomography.
Watanabe T, Song C, Murata K, Kureha T & Suzuki D (2018) Seeded Emulsion Polymerization of Styrene in the Presence of Water-Swollen Hydrogel Microspheres. ACS Lungmuir 34(29), 8571-8580. WEB

Introduces a method for three-dimensional visualization of the chromosome-like DNA structure of cyanobacteria during division using a cryogenic ultra-high voltage electron microscope.
Murata K, & Kaneko Y (2018) Visualization of DNA Compaction in Cyanobacteria by High-voltage Cryo-electron Tomography. J Visual Exper 137, e57197. WEB
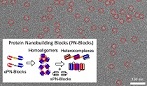
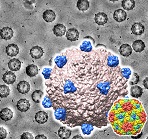
Self-assembled supramolecular nanostructures constructed by protein nanoblocks were observed with an electron microscope.
Kobayashi N, Inano K, Sasahara K, Sato T, Miyazawa K, Fukuma T, Hecht M, Song C, Murata K, & Arai R (2018) Self-Assembling Supramolecular Nanostructures Constructed from de Novo Extender Protein Nanobuilding Blocks. ACS Synth Biol 7, 1381-1394. WEB

The dynamic structural change of the ClpB protein that regenerates the aggregated protein was investigated using an electron microscope.
Uchihashi T, Watanabe Y, Nakazaki Y, Yamasaki T, Watanabe H, Maruno T, Ishii K, Uchiyama S, Song C, Murata K, Iino R, & Ando T (2018) Dynamic Structural States of ClpB Involved in Its Disaggregation Function. Nature Commun 9, 2147. WEB
The morphology of the protozoan parasite was investigated using SBF-SEM. The structure essential for the growth of the protozoa was revealed.
Asare KK, Sakaguchi M, Lucky AB, Asada M, Miyazaki S, Katakai Y, Kawai S, Song S, Murata K, Yahata K, & Kaneko O (2018) The Plasmodium knowlesi MAHRP2 ortholog localizes to structures connecting Sinton Mulligan's clefts in the infected erythrocyte. Parasitol Int 67, 481-492. WEB

A structure of Melbournevirus was investigated by cryo-EM, showing T=304 capsid
Okamoto K, Miyazaki N, Reddy HKN., Hantke MF, Maia FRNC., Larsson DSD, Abergel C, Claverie JM, Hajdu J, Murata K, & Svenda M (2018) Cryo-EM structure of a Marseilleviridae virus particle reveals a large internal microassembly. Virology 516, 239-245. WEB
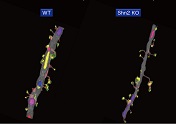
A model of schizophrenia and intellectual disability, "Schnurri-2 knockout mouse" was investigated by SBF-SEM, showing premature morphological features in the hippocampal dentate gyrus of the brain.
Nakao A, Miyazaki N, Ohira K, Hagihara H, Takagi T, Usuda N, Ishii S, Murata K, & Miyakawa T (2017) Immature morphological properties in subcellular-scale structures in the dentate gyrus of Schnurri-2 knockout mice: a model for schizophrenia and intellectual disability. Mol Brain 10:60. WEB

Order of the molecules in chitinase crystal was determined by cryo - electron microscope tomography, which made possible the quantitative measurement of reactive element process of chitinase.
Nakamura A, Tasaki T, Okuni Y, Song C, Murata K, Kozai T, Hara M, Sugimoto H, Suzuki K, Watanabe T, Uchihashi T, Noji H, & Iino R (2017) Rate constants, processivity, and productive binding ratio of chitinase A revealed by single-molecule analysis. Phys Chem ChemvPhys 20(5), 3844. WEB
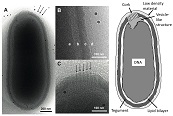
The detailed structure of the largest virus "Pithovirus" was investigated by cryo-electron microscopes.
Okamoto K, Miyazaki N, Song C, Maia FRNC, Reddy HKN, Abergel C, Claverie J-M, Hajdu J, Svenda M, Murata K (2017) Structural variability and complexity of the giant Pithovirus sibericum particle revealed by high-voltage electron cryo-tomography and energy-filtered electron cryo-microscopy. Sci Rep 7, 13291. WEB
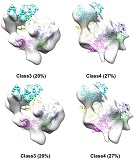
The flexible structure of UGGT, a enzyme for protein quality control, was visualized by cryo-EM.
Satoh T, Song C, Zhu T, Toshimori T, Murata K, Hayashi Y, Kamikubo H, Uchihashi T, & Kato K (2017) Visualisation of a flexible modular structure of the ER folding-sensor enzyme UGGT. Sci Rep 7:12142 WEB
A review of cryo-EM for investigating dynamic protein structures.
Murata K & Wolf M (2017) Cryo-electron microscopy for structural analysis of dynamic biological macromolecules. Biochim Biophys Acta 1862(2), 324-334. WEB
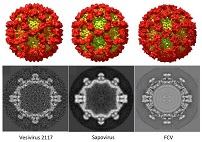
Diversity of structures across genera in the calicivirus family was investigated by cryo-EM.
Conley M, Emmott E, Orton R, Taylor D, Carneiro DG, Murata K, Goodfellow IG, Hansman GS, & Bhella D (2017) Vesivirus 2117 capsids more closely resemble sapovirus and lagovirus particles than other known vesivirus structures. J Gen Virol 98, 68–76. WEB
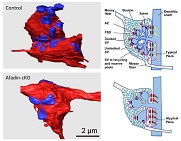
Morphological investigation by SBF-SEM revealed that afadin molecule plays an important role in morphogenesis of mouse hippocampal mossy fiber synapse.
Sai K, Wang S, Kaito A, Fujiwara T, Maruo T, Itoh Y, Miyata M, Sakakibara S, Miyazaki N, Murata K, Yamaguchi Y, Haruta T, Nishioka H, Motojima Y, Komura M, Kimura K, Mandai K, Takai Y, & Mizoguchi A (2017) Multiple roles of afadin in the ultrastructural morphogenesis of mouse hippocampal mossy fiber synapses. J Comp Neurol 525:2719–2734. WEB
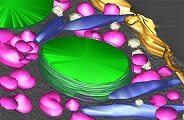
High voltage TEM revealed that the "PV" membrane formed by the symbiotic chlorella with the host 's paramecium is firmly interacting with the host mitochondria via a structure of wedge.
Song C, Murata K, & Suzaki T (2017) Intracellular symbiosis of algae with possible involvement of mitochondrial dynamics. Sci Rep 7, 1221. WEB
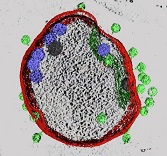
The moment that cyanophage P-SSP7 adsorbed to and infected the host cyanobacteria was visualied by cryo-electron tomography.
Murata K, Zhang Q, Gerardo Galaz-Montoya J, Fu C, Coleman ML, Osburne MS, Schmid MF, Sullivan MB, Chisholm SW, & Chiu W (2017) Visualizing Adsorption of Cyanophage P-SSP7 onto Marine Prochlorococcus. Sci Rep 7, 44176. WEB

Hydrogel microparticles expected to be applied to drug delivery carriers. The hydrated structure was visualized by cryo-EM.
Watanabe T, Kobayashi C, Song C, Murata K, Kureha T, Suzuki D (2016) Impact of Spatial Distribution of Charged Groups in Core Poly( N ‑ isopropylacrylamide)-Based Microgels on the Resultant Composite Structures Prepared by Seeded Emulsion Polymerization of Styrene.Langmuir 32:12760–12773. WEB
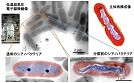
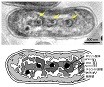
It was found that cyanobacteria produce a structure similar to chromosomes when cell division, and this structure was revealed by high voltage cryo-EM.
Murata K, Hagiwara S, Kimori Y, & Kaneko Y (2016) Ultrastructure of compacted DNA in cyanobacteria by high-voltage cryo-electron tomography.Sci Rep, 6: 34934. WEB
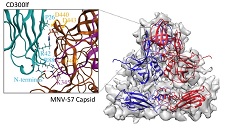
The functional receptor of mouse norovirus was identified as a proteins CD300lf and CD300ld, and the binding site of the receptors were simulated on our cryo-EM map.
Haga K, Fujimoto A, Takai-Todaka R , Miki M, Doan YH, Murakami K, Yokoyama M, Murata K, Nakanishi A, Katayama K. (2016) Functional receptor molecules CD300lf and CD300ld within the CD300 family enable murine noroviruses to infect cells. Proc Natl Acad Sci USA 113(41), E6248–E6255. WEB
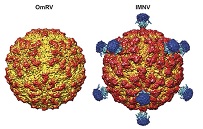
Structure of toti-like virus, Omotoriver virus (OmRV) was investigated by cryo-EM. It was suggested that the long protrusion structure, which was considered necessary for invasion into the host cell from the atmosphere, is not necessary, and the raised structure of the surface of the viral particle directly relates to this.
Okamoto K, Miyazaki N, Larsson DS, Kobayashi D, Svenda M, Mühlig K, Maia FR, Gunn LH, Isawa H, Kobayashi M, Sawabe K, Murata K, Hajdu J. (2016) The infectious particle of insectborne totivirus-like Omono River virus has raised ridges and lacks fibre complexes. Sci Rep, 6: 33170. WEB
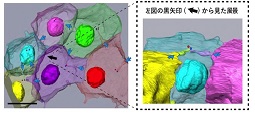
A new mechanism to determine the direction of cell division was found in the ascidian embryo. SBF-SEM observation revealed that the structure extending from the adjacent cell determines the direction of cell division.
Negishi T, Miyazaki N, Murata K, Yasuo H, & Ueno N. (2016) Physical association between a novel plasma-membrane structure and centrosome orients cell division. Elife, 5, e16550. WEB

3D structure of a spiral microorganism discovered in the see with a depth of 1,200 m was investigated by high voltage TEM, showing it contains a small number of organelles as a characteristic of extreme environment.
Yamaguchi M, Yamada H, Higuchi K, Yamamoto Y, Arai S, Murata K, Mori Y, Furukawa H, Uddin MS Chibana H (2016) High-voltage electron microscopy tomography and structome analysis of unique spiral bacteria from the deep sea. Microscopy 65, 363-369. WEB

3D structure of podocytes in the glomerulus of the kidney was reconstructed using SBF-SEM and its detailed formation process was elucidated.
Ichimura K, Kakuta S, Kawasaki Y, Miyaki T, Nonami T, Miyazaki N, Nakao T, Enomoto S, Arai S, Koike M, Murata K, & Sakai T. (2017) Morphological process of podocyte development revealed by block- face scanning electron microscopy. J Cell Sci, 130, 132–142.

Hydrated structure of polystylen microgels prepared by seedied emulsion polymerization of stylene was investigated by cryo-EM, where polystylene particles were located on the surface of the microgel.。
Kobayashi C, Watanabe T, Murata K, Kureha T, & Suzuki D. (2016). Localization of Polystyrene Particles on the Surface of Poly(N-isopropylacrylamide-co-methacrylic acid) Microgels Prepared by Seeded Emulsion Polymerization of Styrene. Langmuir, 32(6): 1429-1439. WEB
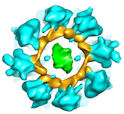
The structure of innexin gap junction ion channel was investigated by ceyo-electron microscopy. It consisted of hexadecameric larger size rings than those of connexin.
Oshima A, Matsuzawa T, Murata K, Tani K, & Fujiyoshi Y. (2016). Hexadecameric structure of an invertebrate gap junction channel. J Mol Biol, 428(6):1227-1236. WEB
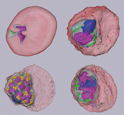
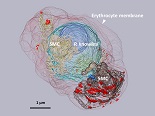
The structual developement in human maralia parasite was investigated by SBF-SEM, suggesting a new mechianism to transport materials between the Erythrocyte membrane and the parasite.
Sakaguchi M, Miyazaki N, Fujioka H, Kaneko O, Murata K.(2016). Three-dimensional analysis of morphological changes in the malaria parasite infected red blood cell by serial block-face scanning electron microscopy. J Struct Biol, 193(3): 162–171. WEB
A detailed structure of legs in tanaid crustacean was investigated using SBF-SEM, wehre spindles were formed from the tip of the legs by mixing materials from two spearate tubes.
Kaji T, Kakui K, Miyazaki N, Murata K, & A R Palmer (2016). Mesoscale morphology at nanoscale resolution: serial block-face scanning electron microscopy reveals fine 3D detail of a novel silk spinneret system in a tube-building tanaid crustacean. Front Zool, 13:14. WEB

The relationship between microtubles (MTs) and microfilaments (MFs) was revealed in plant cells using electron tomgoraphy. MFs make a croslink between MTs forming a bundle of MTs durint the formation of pre-proface bands.
Takeuchi M, Karahara I, Kajimura N, Takaoka A, Murata K, Misaki K, Yonemura S, Staehelin LA & Mineyuki Y. (2016). Single microfilaments mediate the early steps of microtubule bundling during preprophase band formation in onion cotyledon epidermal cells. Mol Biol Cell, 27(11):1809-1820. WEB
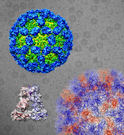
The fist 3D structure of sapovirus revealed by cryo-EM showed a large gap between outer and inner capsid layers. A lot of possible epitops were identified in the inner capsid layer.
Miyazaki N, Taylor D W, Hansman G S, & Murata K. (2015). Antigenic and cryo-electron microscopy structure analysis of a chimeric sapovirus capsid. J Virol, 90(5): 2664-2675. WEB
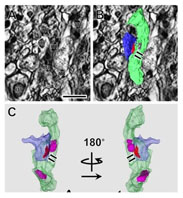
A new method for HVEM tomography was developed to analyze the structure of synapse connections in neural network, where immuno-histochemistry was combined with the OTO en bloc staining. The method makes a possible to find and analyze the structure of the taeget synapse efficiently.
Satoh K, Takanami K, Murata K, Kawata M, Sakamoto T, & Sakamoto H. (2015). Effective synaptome analysis of itch-mediating neurons in the spinal cord : A novel immunohistochemical methodology using high-voltage electron microscopy. Neurosci Let 599 : 86–91. WEB

A mutant of budding yeast lakcking Cdc48p ATPase showing unusual mitochondrial morphology was investigated using SBF-SEM. The mitochondria showed flagmentation and aggregation in the mutant.
Miyazaki N, Esaki M, Ogura T, Murata K. (2014). Serial block-face scanning electron microscopy for three-dimensional analysis of morphological changes in mitochondria regulated by Cdc48p/p97 ATPase. J Struct Biol, 187: 187-93. WEB
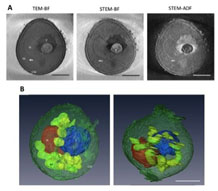
A whole cell structure of a budding yeast was investigated using high-voltage electron tomography. Especially, the bright field STEM revealed the clear 3D structures and their localizations of major organella, nuleus, mitochondoria, valuoles in the cell.
Murata K, Esaki M, Ogura T, Arai S, Yamamoto Y, & Tanaka N. (2014). Whole-Cell Imaging of the Budding Yeast Saccharomyces cerevisiae by High-Voltage Scanning Transmission Electron Tomography. Ultramicroscopy, 146: 39-45. WEB
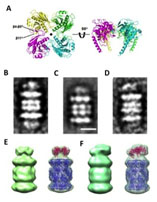
A homolog of the self-assemby factor in 20S protease functions as the activation factor in Archaea. Electron tomography revealed the binding feature of the homolog with 20S protease.
Kumoi K, Satoh T, Murata K, Hiromoto T, Mizushima T, Kamiya Y, Noda M, Uchiyama S, Yagi H & Kato K. (2013). An archaeal homolog of proteasome assembly factor functions as a proteasome activator. PloS one 8: e60294. WEB
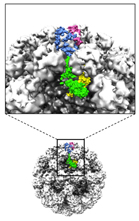
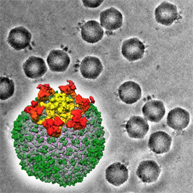
The binding site of monoclonal aintibody recognizing the capsid domain inside of the norovirus capsid was identified.
Hansman G, Taylor D, McLellan J, Smith T, Georgiev I, Tame J, Park SY, Yamazaki M, Gondaira F, Miki M, Katayama K, Murata K* & Kwong P*. (2012) Structural basis for broad detection of genogroup II noroviruses by a monoclonal antibody that binds to a site occluded in the viral particle. J Virol 86: 3635-3646. *Corresponding authors WEB
Phase plate electron microscopy makes more easy to determine protein structures at nano- and subnanometer resolutions.
Murata K, Liu X, Danev R, Jakana J, Schmid MF, King J, Nagayama K, & Chiu W (2010) Zernike Phase Contrast Cryo-Electron Microscopy and Tomography for Structure Determination at Nanometer and Sub-Nanometer Resolutions. Structure 18(8) : 903-912. WEB

Structure of alpha1-beta complex in Dihydropyridine Receptor (DHPR: L-type voltage-gated Ca2+ channel) was reconstructed using single particle electron microscopy, which suggests the localization of five subunits in DHPR.
Murata K, Nishimura S, Kuniyasu A, & Nakayama H (2010) Three-dimensional structure of the alpha1-beta complex in the skeletal muscle dihydropyridine receptor by single-particle electron microscopy. J Electron Microsc 59(3) : 215-226. WEB

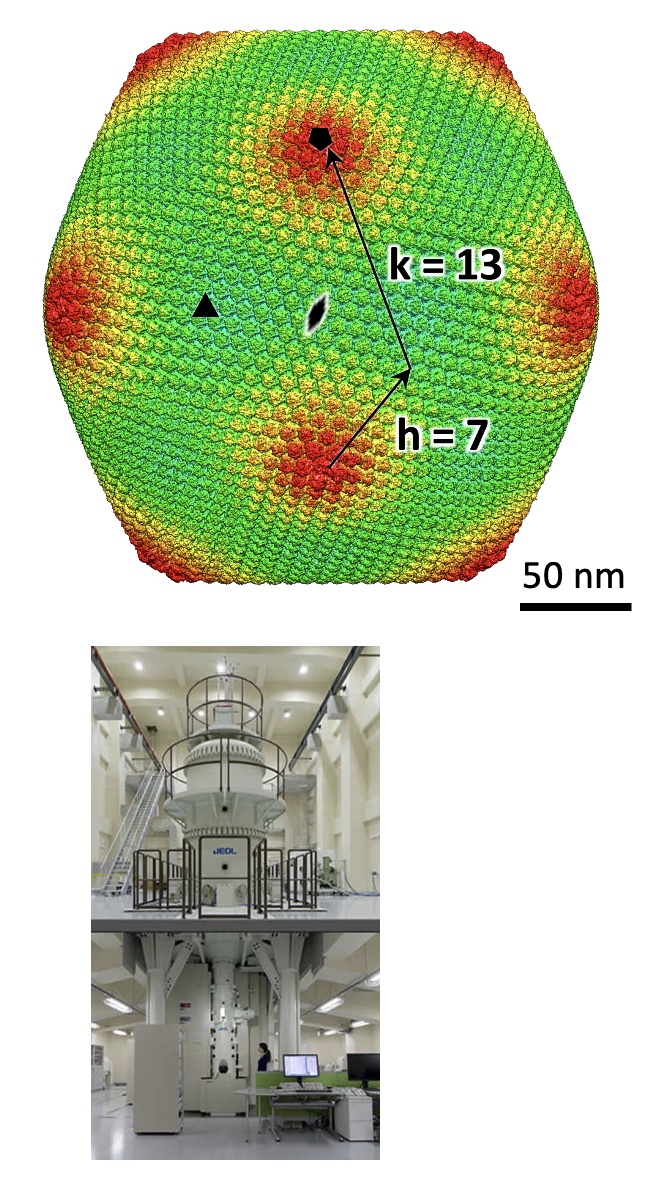
The high-end TITAN Krios G4 (TFS) having a 300kV cold-field emission gun and a pos- column energy filter (Selectris X) is used for single particle anaysis, electron tomography, and micrED.
Gun: 300kV C-FEG
Energy filter: Post column Selectris X
Camera: Falcon4
Software: EPU

The electron microscope JEM2200FS (JEOL) having a 200kV Thermal Schottky and an in-column energy filter, is used for grid screening and training for cryo-EM.
Gun: 200kV Thermal Schottky FEG
Specimen holder: Gatan 622 cryo-specimen holder
Energy filter: In-column Omega-type
Camera: DE-20

Cryo-FIB-SEM, Aquilos2 (TFS) is used for producing a thin cryo-section of biological samples. An additional fluoescence microscope unit (iFLM) makes it easy to find the target area .
SEM: Thermal Schottky FEG
FIB: Ga Liquid metal ion source
CLEM: iFLM
Our lab. belongs to Department of Physiological Sciences,
School of Life Science, The Graduate University for
Advanced Studies (SOKENDAI).
Copyright (C) 2021 NINS Murata Lab. All Right Reserved