We investigate structures of biomolecules, biomacromolecular complexes, and cellular architecture using electron microscopy, and reveal a relationship between the structure and the function.

We have succeeded in elucidating the fiber structure of the familial Tottori type (D7N mutation) of amyloid beta (Aβ), the causative agent of Alzheimer's disease, through experiments utilizing the microgravity environment of the International Space Station.
Yagi-Utsumi M, Yanaka S, Burton-Smith RN, Song C, Ganser C, Yamazaki C, Kasahara H, Shimazu T, Uchihashi T, Murata K, Kato K (2025) Microgravity-Assisted Exploration of the Conformational Space of Amyloid β Affected by Tottori-Type Familial Mutation D7N. ACS Chem Neurosci 16(14) 2682-2690. WEB
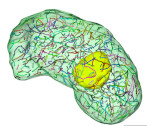
We used volume EM to perform a detailed analysis of the nuclear structure of the dinoflagellate Oxyris marina, revealing for the first time that this species has nearly 400 chromosomes (dinokaryons) unique to this species.
Fukuda Y, Suzaki T, Murata K, Chihong S (2025) Novel ultrastructural features of the nucleus of the ancestral dinoflagellate Oxyrrhis marina as revealed by freeze substitution fixation. Front Protistol 3 1512258. WEB
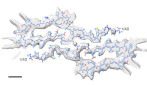
Cryo-EM structural analysis of wild-type Aβ(1-40) fibers formed on Earth in a manner similar to that in the microgravity environment of the ISS revealed a novel J-shaped structure.
Burton-Smith RN, Yagi-Utsumi M, Yanaka S, Song C, Murata K, Kato K (2025) Elucidating the Unique J-Shaped Protomer Structure of Amyloid-β(1-40) Fibril with Cryo-Electron Microscopy. Int J Mol Sci 26(3) 1179. WEB
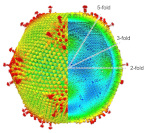
The capsid structure of Medusavirus was analyzed at 7 Å resolution using cryo-EM, revealing structural changes in the capsid that accompany the particle formation process.
Watanabe R, Song C, Takemura T, Murata K* (2024) Subnanometer structure of medusavirus capsid during maturation using cryo-electron microscopy. J Virol 98(9), e0043624. WEB
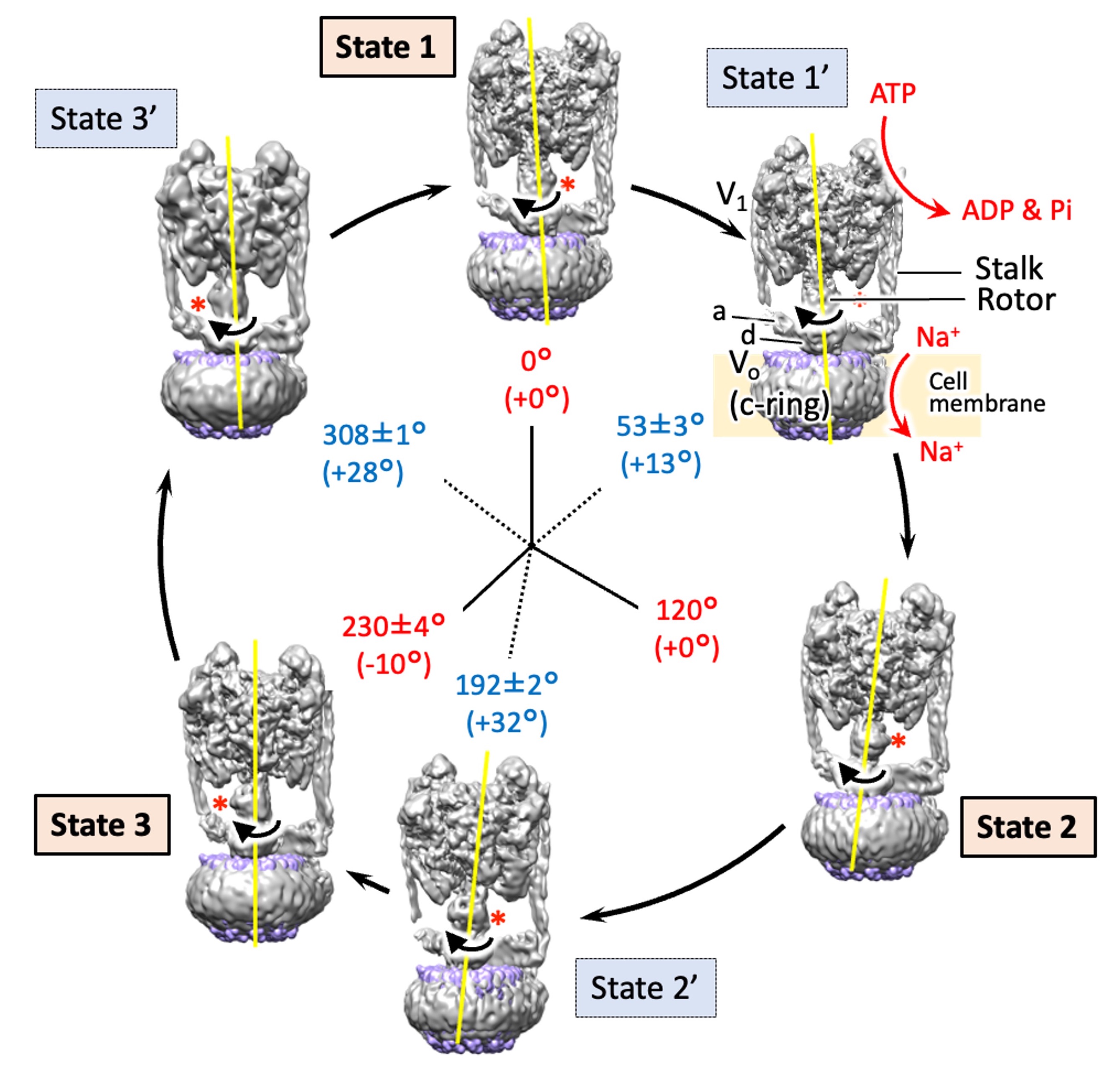
The structures of six states of a rotary sodium ion pump are revealed using cryo-electron microscopy,elucidating the branched mechanism by which sodium is transported by hydrolyzing ATP. .
Raymond N. Burton Smith, Chihong Song, Hiroshi Ueno, Takeshi Murata, Ryota Iino, Kazuyoshi Murata* (2023) Six states of Enterococcus hirae V-type ATPase reveals non-uniform rotor rotation during turnover. Comm Bio 6 755. WEB
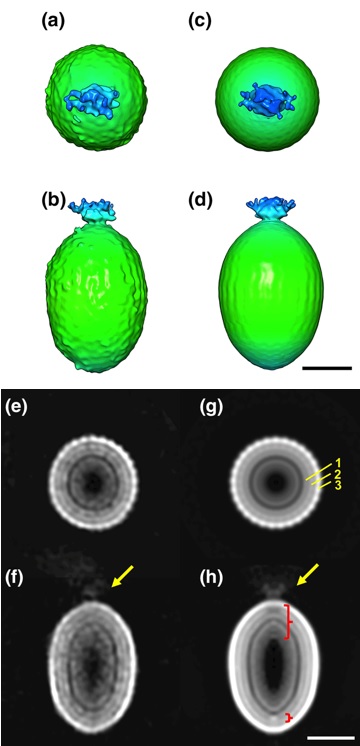
The detailed particle structure of Tanaivirus was revealed using cryo-electron microscopy, revealing the dynamic structural changes of the capsid that occur during infection.
Kenta Okamoto*, Chihong Song, Han Wang, Miako Sakaguchi, Christina Chalkiadaki, Naoyuki MIyazaki, Takeshi Nabeshima, Kouichi Morita*, Shingo Inoue, and Kazuyoshi Murata (2023) Structure and its transformation of elliptical nege-like virus Tanay virus. J Gen Vir 104(6),001863. WEB
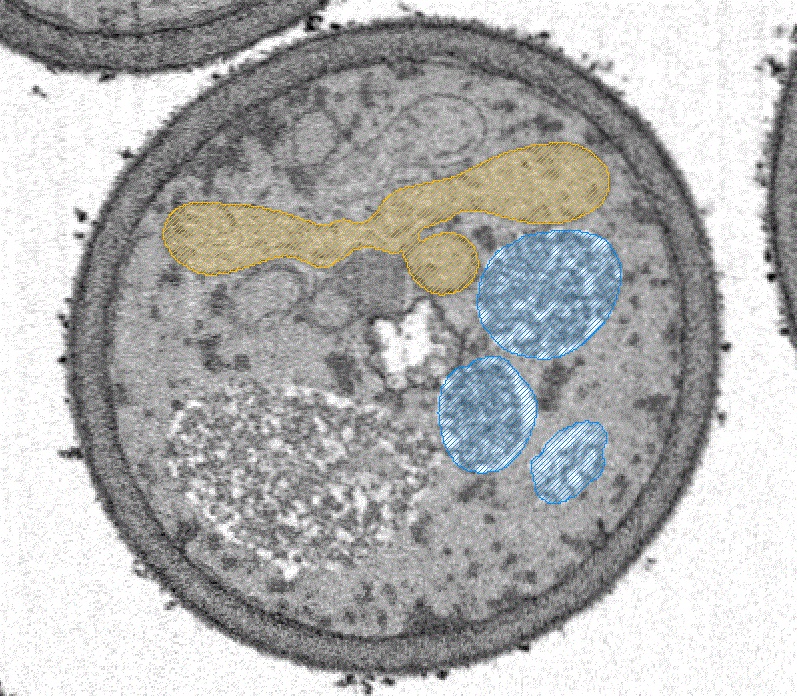
New factor of mitochondrial fission in mitophagy was discovered and the process was visualized by 3D-SEM.
Tomoyuki Fukuda#, Kentaro Furukawa#, Tatsuro Maruyama#, Shun-Ichi Yamashita, Daisuke Noshiro, Chihong Song, Yuta Ogasawara, Kentaro Okuyama, Jahangir Md. Alam, Manabu Hayatsu, Tetsu Saigusa, Keiichi Inoue, Kazuho Ikeda, Akira Takai, Lin Chen, Vikramjit Lahiri, Yasushi Okada, Shinsuke Shibata, Kazuyoshi Murata, Daniel J. Klionsky, Nobuo N. Noda*, Tomotake Kanki* (2023) The mitochondrial intermembrane space protein mitofissin drives mitochondrial fission required for mitophagy. Mol Cell 83(12),2045-2058. WEB
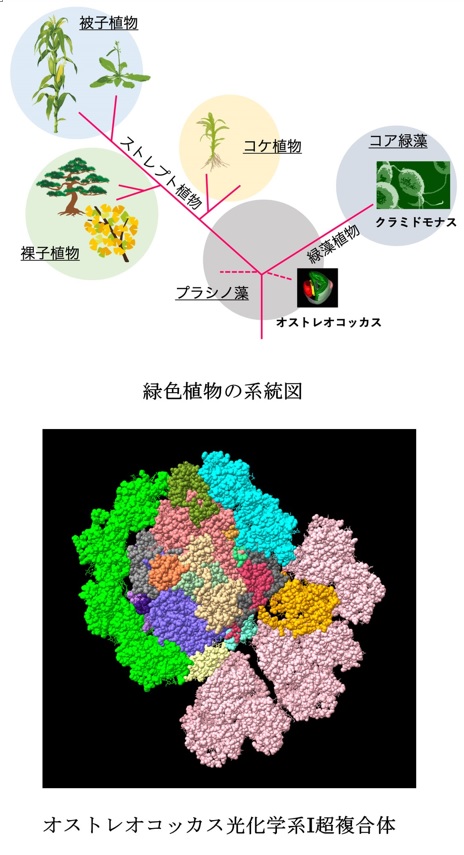
The unique structure of acient type PSI in Ostreococcal was revealed using cryo-electron microscopy.
Asako Ishii, Jianyu Shan, Xin Sheng, Eunchul Kim, Akimasa Watanabe, Makio Yokono, Chiyo Noda, Chihong Song, Kazuyoshi Murata, Zhenfeng Liu, Jun Minagawa (2023) The photosystem I supercomplex from a primordial green alga Ostreococcus tauri harbors three light-harvesting complex trimers. eLife 84488. WEB
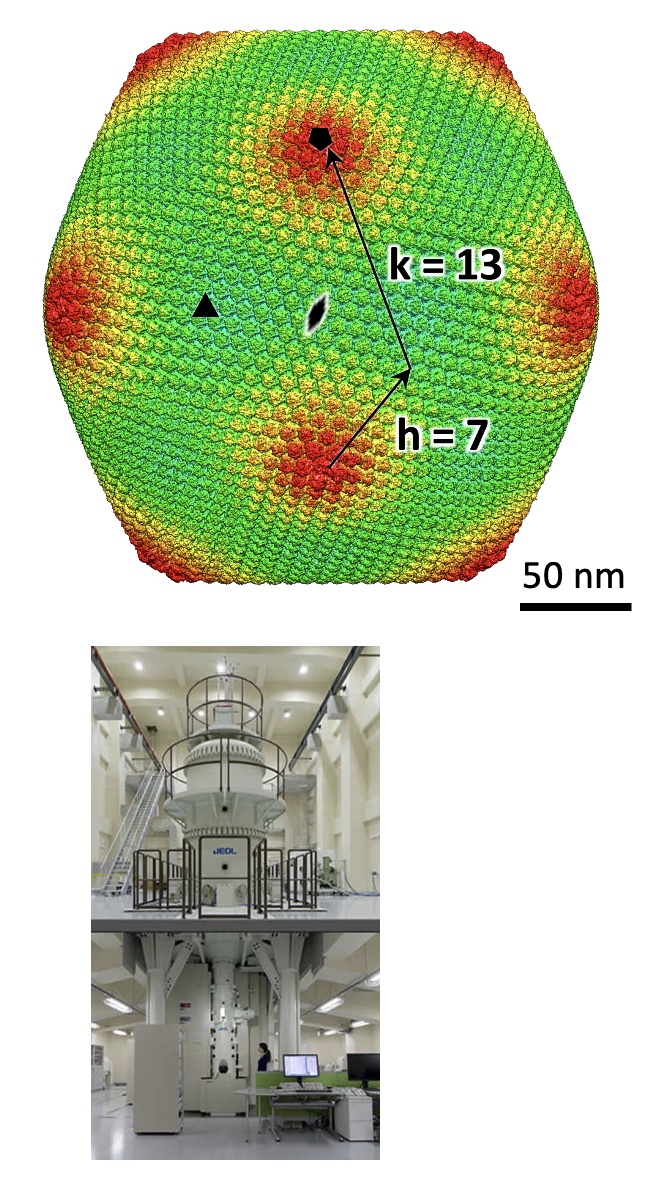
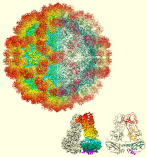
Structure of a giantvirus, tokyovirus, was revealed at 7.7Å resolution by high-voltage cryo-EM single particle analysis.
Chihara A, Burton-Smith RN, Kajimura N, Mitsuoka K, Okamoto K, Song C, Murata K (2022) A novel capsid protein network allows the characteristic inner membrane structure of Marseilleviridae giant viruses. Sci Rep 12(1):21428. WEB
Hydrated structures of nano-composite microgels produced with different components were revealed by cryo-electron tomography.
Nishizawa Y, Watanabe T, Noguchi T, Takizawa M, Song C, Murata K, Minato H, Suzuki D (2022) Durable gelfoams stabilized by compressible nanocomposite microgels. Chem Commun (Camb) 58, 12927-12930. WEB
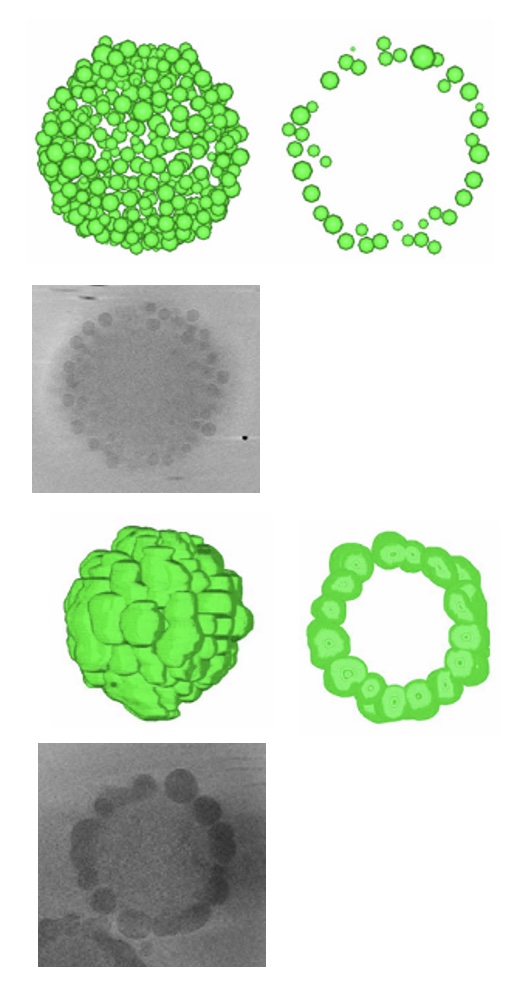
The structure of the sapovirus shell (capsid), one of the causative viruses of mass diarrhea, was clarified for the first time in the world at the atomic level.
Miyazaki N, Song C, Oka T, Miki M, Murakami K, Iwasaki K, Katayama K, Murata K* (2022) Atomic structure of human sapovirus capsid by single particle cryo-electron microscopy revealed a unique capsid protein conformation in caliciviruses. J Virol 96(9), e0029822. WEB
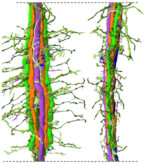
The neural circuit (connectomics) of the brain region where the light information entering from the compound eye of the swallowtail butterfly is initially processed has been clarified.
Matsushita A, Stewart F, Ilić M, Chen PJ, Wakita D, Miyazaki N, Murata K, Kinoshita M, Belusic G, Arikawa K (2022) Connectome of the lamina reveals the circuit for early color processing in the visual pathway of a butterfly. Cur biology 32(10):2291.e3, 00506-1. WEB
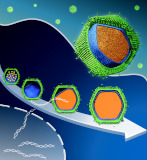
Medusavirus, a giant virus, shows a unique maturation process in host amoeba.
Watanabe R, Song C, Kayama Y, Takemura M, Murata K* (2022) Particle morphology of Medusavirus inside and outside cells reveals new maturation process of giant virus. J Viol 96(7), e0185321. WEB

The high-end TITAN Krios G4 (TFS) having a 300kV cold-field emission gun and a pos- column energy filter (Selectris X) is used for single particle anaysis, electron tomography, and micrED.
Gun: 300kV C-FEG
Energy filter: Post column Selectris X
Camera: Falcon4
Software: EPU

The electron microscope JEM2200FS (JEOL) having a 200kV Thermal Schottky and an in-column energy filter, is used for grid screening and training for cryo-EM.
Gun: 200kV Thermal Schottky FEG
Specimen holder: Gatan 622 cryo-specimen holder
Energy filter: In-column Omega-type
Camera: DE-20

Cryo-FIB-SEM, Aquilos2 (TFS) is used for producing a thin cryo-section of biological samples. An additional fluoescence microscope unit (iFLM) makes it easy to find the target area .
SEM: Thermal Schottky FEG
FIB: Ga Liquid metal ion source
CLEM: iFLM
Our lab. belongs to Department of Physiological Sciences,
School of Life Science, The Graduate University for
Advanced Studies (SOKENDAI).
Copyright (C) 2021 NINS Murata Lab. All Right Reserved