Key Points
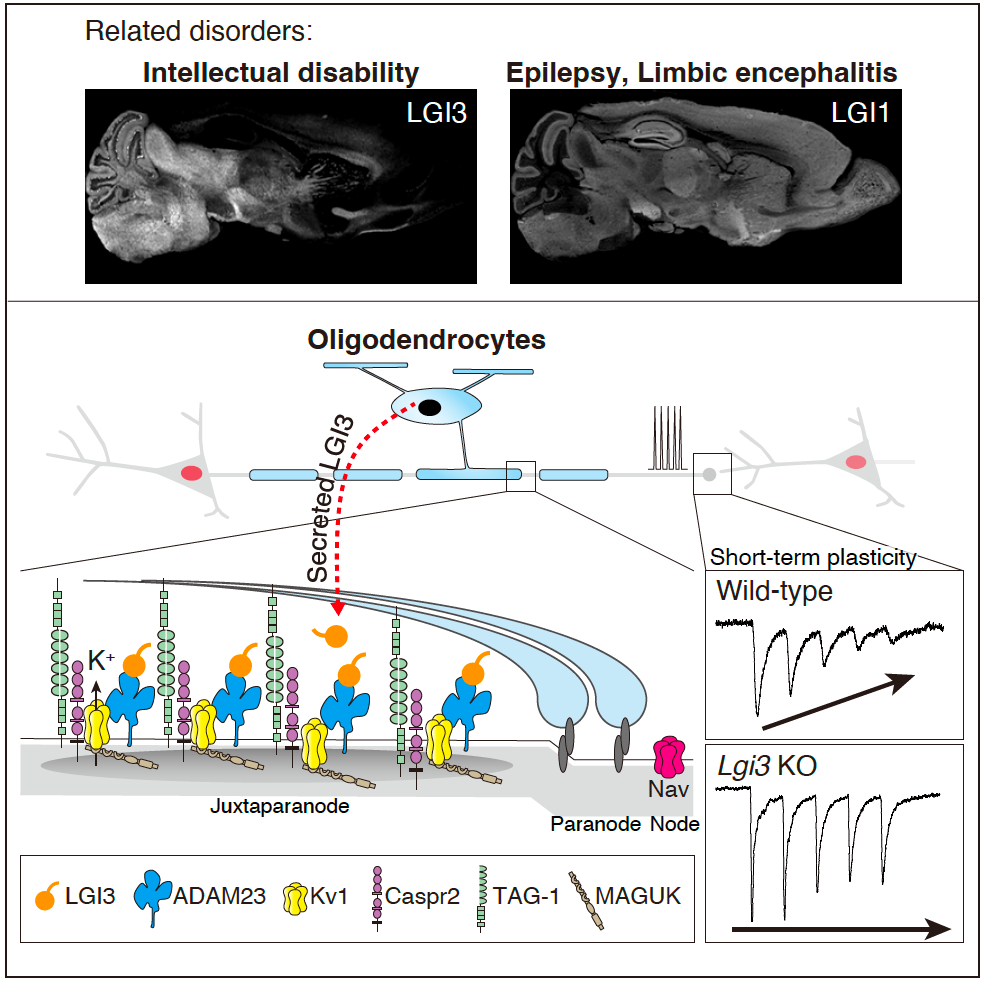
• LGI3 is a recently recognized intellectual disability (ID)-associated gene, however, its pathogenetic mechanism has been unclear.
• LGI3 is secreted from oligodendrocytes and regulates Kv1 channel clustering at the juxtaparanodes through the interaction with its receptor, ADAM23.
• Loss of LGI3 results in abnormal action potential conductions and synaptic transmission which may be the pathogenic mechanisms of LGI3-related ID.
• These findings would provide insights into the new therapeutic strategies for the LGI3- and Kv1 channel-related neurological disorders.
|
A research group led by Drs. Yuri Miyazaki, Yuko Fukata, and Masaki Fukata (Division of Neuropharmacology, Nagoya University Graduate School of Medicine) in collaboration with Drs. Takeshi Otsuka, Masumi Hirabayashi et al. (National Institute for Physiological Sciences, National Institutes of Natural Sciences) reported that an intellectual disability (ID)-related protein LGI3 is secreted from oligodendrocytes and regulates voltage-dependent potassium channel (Kv1) at the juxtaparanodes of myelinated axons through the interaction with a receptor protein, ADAM23. Loss of LGI3 causes disrupted localization of ADAM23 and Kv1 channels at the juxtaparanodes resulting in altered action potential waveforms and short-term synaptic plasticity in mice. We propose a molecular pathway, juxtaparanodal LGI3–ADAM23–Kv1 channel, for understanding neurodevelopmental disorders including ID. |
Research Background
Functional characterization of disease-associated genes is vital for understanding physio-pathological brain functions. LGI3 is a member of the LGI family genes (LGI1-LGI4) and has recently been reported as an ID-related gene, however, its pathophysiological mechanism has been unclear.
In this study, we find that LGI3 is uniquely secreted from oligodendrocytes in the brain and enriched at juxtaparanodes of myelinated axons. Proteomic analysis using epitope-tagged LGI3 knock-in mice shows that LGI3 uses ADAM23 as a receptor and selectively co-assembles with Kv1 channels. A lack of LGI3 in mice disrupts the juxtaparanodal clustering of ADAM23 and Kv1 channels and suppresses Kv1 channel-mediated short-term synaptic plasticity. Given the loss of function phenotypes of the LGI3, we propose a molecular pathway, juxtaparanodal LGI3–ADAM23–Kv1 channel, for understanding neurodevelopmental disorders.
Research Results
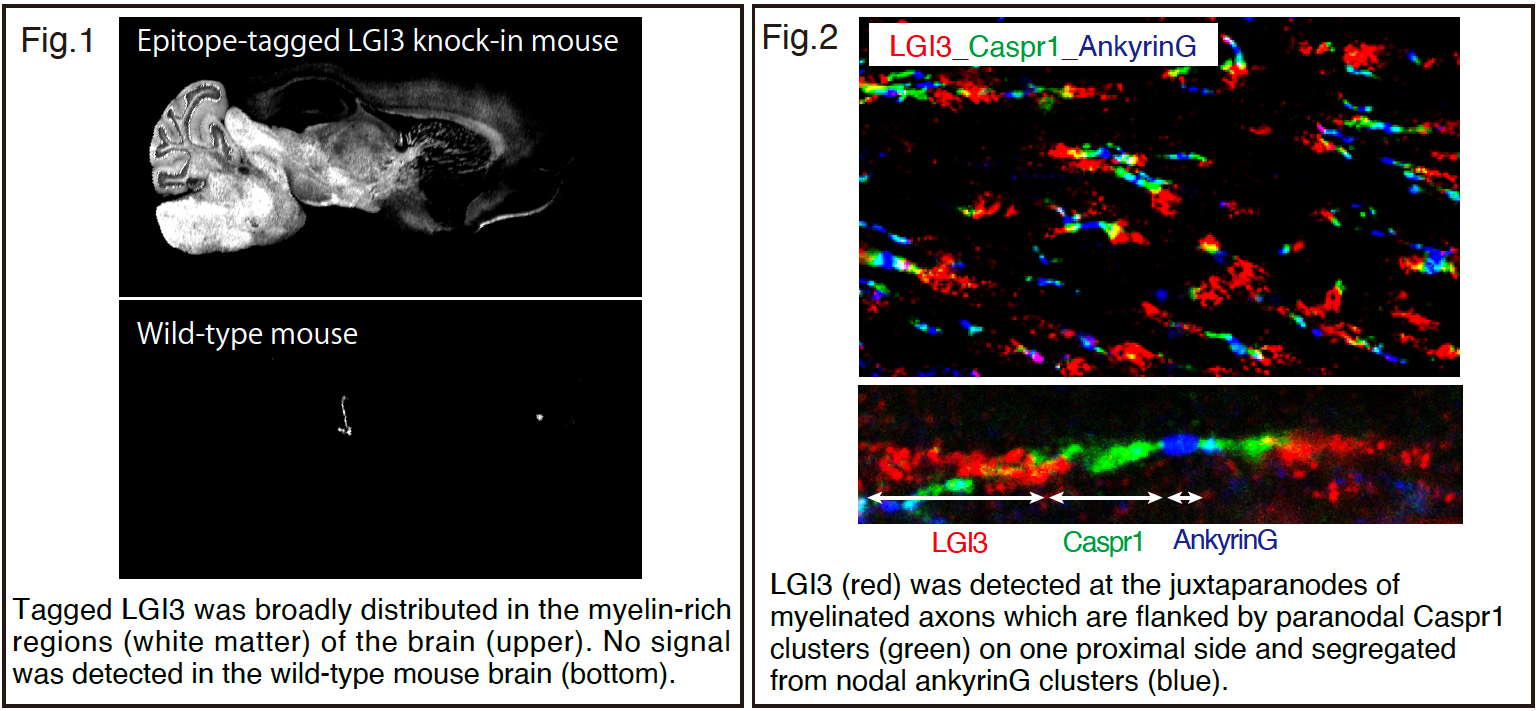
First, we generated the epitope-tagged LGI3 knock-in mouse and investigated the brain subregional distribution. LGI3 was broadly distributed in the myelin-rich regions of the brain such as deeper layers of the cortex, corpus callosum, internal capsule, thalamus, midbrain, medulla, and cerebellar white matter (Figure 1). Then, we examined the localization of LGI3 along myelinated axons and found that LGI3 was localized in the juxtaparanode (Figure 2). We also demonstrated that LGI3 was predominantly secreted from oligodendrocytes by using mouse genetics approach.
To comprehensively identify the LGI3-associated protein complexes in the brain, we purified LGI3 from the mouse brains and performed shot-gun proteomic analysis. We identified ADAM23 family proteins (known as receptors for other LGI proteins) and Kv1 channels as the main LGI3-interacting proteins.
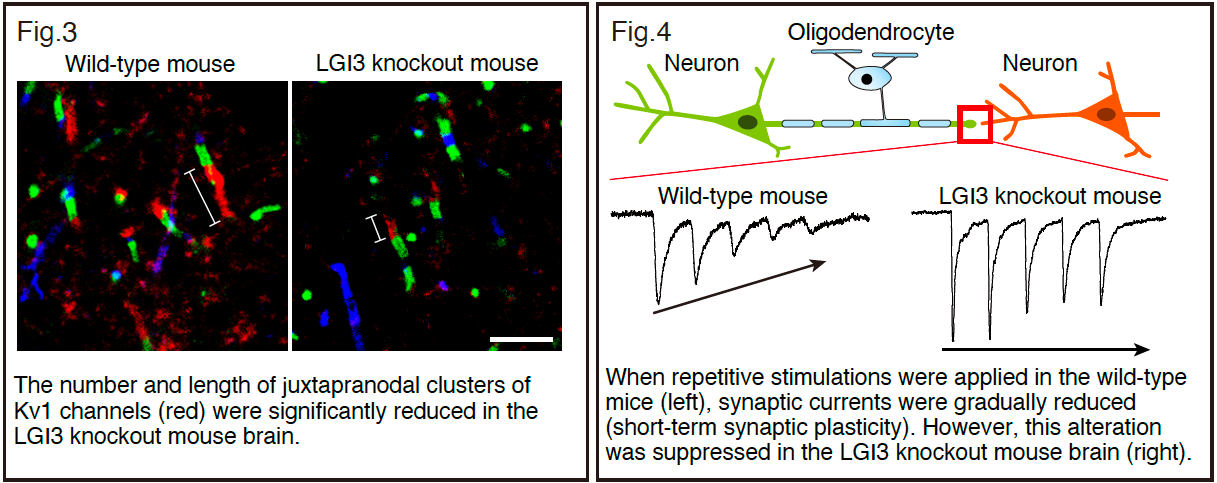
Next, to investigate whether and how LGI3 physiologically functions with the associated proteins, we generated LGI3 knockout mice. The immunohistochemical analysis showed that the clustering of ADAM23 and Kv1 channels at the juxtaparanodes was significantly disrupted (Figure 3). In addition, electrophysiological analysis revealed that action potential waveforms and short-term synaptic plasticity were altered in the LGI3 knockout mouse brains in a Kv1 channel-dependent manner (Figure 4).
These findings strongly suggest that the altered electrophysiological properties caused by LGI3 dysfunction (mutation) are the pathophysiological mechanisms of LGI3-related ID.
Research Summary and Future Perspective
This study demonstrated that ID-related LGI3 protein and its receptor ADAM23 essentially regulates the juxtaparanodal Kv1 channel clustering and thereby controls short-term synaptic plasticity in the brain. We propose “juxtaparanodal LGI3–ADAM23–Kv1 channel” as a potential functional pathway affected in neurodevelopmental disorders such as ID. It has been reported that other LGI family proteins are localized at the different subcellular regions (e.g., synapses and axon initial segment), and some parts of them may function with Kv1 channels. We will investigate the mode of interaction/action of LGI family–ADAM23 family–Kv1 channels for their physiological roles in the brain. Clarifying the whole picture of LGI–ADAM family-related neurological/brain disorders will contribute to discovering additional molecular pathways for brain functioning and therapeutic strategies.
Publication
Journal: Cell Reports
Title: Oligodendrocyte-derived LGI3 and its receptor ADAM23 organize juxtaparanodal Kv1 channel clustering for short-term synaptic plasticity
Authors: Yuri Miyazaki,
1,2 Takeshi Otsuka,
3,4 Yoko Yamagata,
5 Toshihiro Endo,
6 Makoto Sanbo,
7 Hiromi Sano,
8 Kenta Kobayashi,
4,9 Hiroki Inahashi,
2 Hans-Christian Kornau,
10,11 Dietmar Schmitz,
10,11 Harald Prüss,
10,12,13 Dies Meijer,
14,15 Masumi Hirabayashi,
4,7 Yuko Fukata,
2,16* Masaki Fukata
1,2,4*
1. Division of Neuropharmacology, Nagoya University Graduate School of Medicine, Nagoya 466-8550, Japan.
2. Division of Membrane Physiology, Department of Molecular and Cellular Physiology, National Institute for Physiological Sciences, National Institutes of Natural Sciences, Okazaki, Aichi 444-8787, Japan.
3. Section of Cellular Electrophysiology, National Institute for Physiological Sciences, National Institutes of Natural Sciences, Okazaki, Aichi 444-8787, Japan.
4. Graduate Institute for Advanced Studies, SOKENDAI, Okazaki, Aichi 444-8585, Japan.
5. Section of Multilayer Physiology, National Institute for Physiological Sciences, National Institutes of Natural Sciences, Okazaki, Aichi 444-8585, Japan.
6. Phenovance LLC, Kashiwa, Chiba 277-0882, Japan.
7. Section of Mammalian Transgenesis, Center for Genetic Analysis of Behavior, National Institute for Physiological Sciences, National Institutes of Natural Sciences, Okazaki, Aichi 444-8787, Japan.
8. Division of Behavioral Neuropharmacology, International Center for Brain Science, Fujita Health University, Toyoake, Aichi 470-1192, Japan.
9. Section of Viral Vector Development, Center for Genetic Analysis of Behavior, National Institute for Physiological Sciences, National Institutes of Natural Sciences, Okazaki, Aichi, 444-8585, Japan.
10. German Center for Neurodegenerative Diseases (DZNE) Berlin, Berlin, Germany.
11. Neuroscience Research Center (NWFZ), Cluster NeuroCure, Charité-Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Berlin, Germany.
12. Helmholtz Innovation Lab BaoBab (Brain antibody-omics and B-cell Lab), Berlin, Germany.
13. Department of Neurology and Experimental Neurology, Charité-Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Berlin, Germany.
14. Centre for Discovery Brain Sciences, University of Edinburgh, Edinburgh. UK.
15. Muir Maxwell Epilepsy Centre, University of Edinburgh, Edinburgh, UK.
16. Division of Molecular and Cellular Pharmacology, Nagoya University Graduate School of Medicine, Nagoya 466-8550, Japan.
*Correspondence
DOI: 10.1016/j.celrep.2023.113634